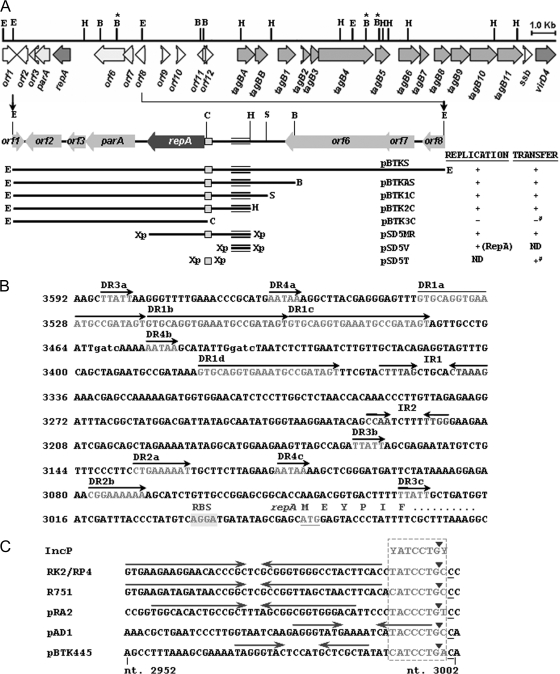
FIG. 2.
(A) Identification of the minimal-replicon, oriV, and oriT regions of pBTK445. The 24,440-bp sequenced region of pBTK445 is drawn to scale as a thick horizontal line. ORFs are represented by arrows, with arrowheads indicating the transcriptional orientation. Vertical lines above the fragment represent the restriction sites (B, BamHI; B marked with an asterisk, BglII; C, ClaI; E, EcoRI; H, HindIII; S, SacII; Xp, XbaI), derived from the corresponding primers. The EcoRI and HindIII sites are used for the construction of banks, and the BglII sites are used for knockout mutants (see Materials and Methods). The 5.7-kb EcoRI fragment used for the characterization of the replication region is diagramed below. Restriction fragments incorporated into different constructs are indicated as horizontal lines, with the enzyme abbreviations at their two ends. In the “Replication” column, a plus sign indicates the ability of a plasmid to replicate in T. kashmirensis, and a minus sign indicates the inability to do so. RepA protein, when supplied in trans, is shown in parentheses. ND, not determined. The 264-bp minimal oriV is represented as triple horizontal lines. The mobilization abilities of the derivatives are given under the “Transfer” column. A plus sign indicates a mobilization frequency of >10−5, and a minus sign indicates a mobilization frequency of <10−7. Conjugative mating was performed with T. kashmirensis strains as the recipients. Experiments using E. coli as the recipient are marked with a pound sign. The 100-bp minimal oriT is represented by squares in the plasmid diagrams. Inferences regarding the replication and conjugation abilities of the constructs are drawn from three different experiments. (B) Organization of the repA gene and the corresponding cis-acting elements. The reverse complementary sequence of the region is shown (nt 2953 to 3592). DR and IR, direct and inverted repeats, respectively. Dam methylase recognition sites are represented by lowercase letters (gatc). (C) DNA sequence comparison of the oriT regions of RP4/RK2, R751, pRA2, pAD1, and pBTK445. The IncP-like oriT consensus sequence is boxed. Inverted repeats are marked by arrows. Solid arrowheads indicate the known or predicted nic (nick) sites.