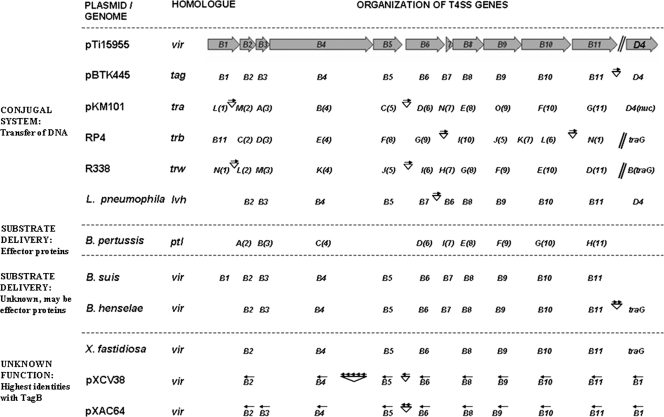
FIG. 4.
Comparison of the T4SS homologs. The virB locus of the A. tumefaciens plasmid pTi15955 (X06826; numbers in parentheses herein refer to GenBank accession numbers), used as the reference, is diagramed at the top. Filled arrows represent genes, with arrowheads indicating their transcriptional orientations. The T4SS homologs are divided into four groups, separated by dashed lines. The presence of a particular homolog in a system is indicated by the name of the gene below the arrow for the corresponding reference gene. Unless otherwise indicated, the genes are transcribed in the same direction as those in pTi15955. Some systems are disrupted by one or many unrelated ORFs (represented by arrows above inverted triangles, with each arrow representing one ORF and the arrowheads indicating their orientations). The tagB genes of pBTK445 are included in the first group along with homologs from plasmids RP4 (M93696 and CAA38334) and R388 (X81123 and X63150), in addition to the genomic counterpart of Legionella pneumophila (Y19029). This group corresponds to systems known to transfer DNA between bacteria. The B. pertussis (BX640422) ptl system, known to deliver the pertussis toxin (substrate) to mammalian cells, is in the second group. The third group consists of systems whose substrates are presently unknown but are postulated to be effector proteins. These systems are represented by the genomic homologs of B. suis (AF141604) and B. henselae (BX897699). The last group consists of systems that have the highest identities with TagB and are available in the database without any functional information. These include the virB loci from the genome of X. fastidiosa (NZ_AAAM0300005), plasmid pXCV38 (AM039950) of Xanthomonas campestris pv. vesicatoria, and plasmid pXAC64 (AE008925) of Xanthomonas axonopodis pv. citri.