Abstract
We have evaluated Photorhabdus insect-related protein (Pir) from Photorhabdus asymbiotica against dengue vectors. PirAB shows larvicidal activity against both Aedes aegypti and Aedes albopictus larvae but did not affect the Mesocyclops thermocyclopoides predator. PirAB expressed the strongest toxicity compared to PirA, PirB, or the mixture of PirA plus PirB. Whether the presence of an enterobacterial repetitive intergenic consensus sequence in PirAB, but not in PirA, PirB, or the mixture of PirA plus PirB, has any impact on biological control efficacy needs further investigation.
Dengue fever is a widespread vector-borne disease and an important global public health problem. Aedes aegypti is the principal vector of dengue virus, the mosquito-borne pathogen that causes dengue fever (9). An effective vaccine against the dengue virus is not yet available; therefore, controlling the Aedes vector is currently the only way to reduce viral transmission. The mosquito larvae have been controlled by biological control toxins, such as those from Bacillus thuringiensis and Bacillus sphaericus. Nevertheless, over the years there has been a gradual development of insect resistance against B. thuringiensis toxins (3). Previous studies found that the PirAB toxin of Photorhabdus luminescens when expressed in Escherichia coli had the ability to kill larvae of Galleria mellonella (11). Photorhabdus spp. are non-free-living Enterobacteriaceae (1, 7). All three species of Photorhabdus have a symbiotic relationship with nematodes of the genus Heterorhabditis (8). Photorhabdus bacteria produce many toxins and other potential virulence factors, and no resistance against the bacteria in any insect host has been reported to date (3, 4). Species of the genus Photorhabdus are pathogenic to most insects when released into the hemolymph (2).
The genome of the insect pathogen P. luminescens subsp. laumondii strain TT01 contains numerous genes predicting toxins, hemolysins, and proteases, which may be important for insect pathogenicity (6). The proteins encoded by the loci plu4093 to plu4092 and plu4437 to plu4436 within P. luminescens TT01, termed PirA and -B, respectively, for “Photorhabdus insect-related proteins A and B,” show similarity to both δ-endotoxins from B. thuringiensis and a developmentally regulated protein from a beetle, Leptinotarsa decemlineata (11). Previously, Photorhabdus asymbiotica had only been isolated from human clinical specimens from the United States and Australia (12). Both wound and blood cultures of these clinical specimens could grow P. asymbiotica, an enteric gram-negative rod that was initially misidentified by the hospital's rapid identification system. Recently, however, this species has also been confirmed to exist in an entomopathogenic symbiosis with a Heterorhabditis nematode in the soil (8).
In this study, larvicidal activity or the ability to kill A. aegypti larvae of P. asymbiotica has been evaluated. The sequence of P. asymbiotica Pir loci was compared with that of P. luminescens, and the toxicity of Pir proteins was assessed via oral biological assay.
The mosquito larvae (A. aegypti and A. albopictus) were reared in dechlorinated tap water in the insectary at the Center of Excellence for Vectors and Vector-Borne Diseases, Faculty of Science, Mahidol University, at 25 to 27°C and 75% relative humidity. Commercial powdered hamster food (0.1 g) was supplied daily. Mesocyclops thermocyclopoides, a crustacean biological control agent of mosquito larvae, was reared in the boiled straw water at the same humidity and temperature and was supplied with paramecium as a daily food.
Mosquitoes were bioassayed with recombinant PirA, PirB, and PirAB proteins orally. Cells of recombinant E. coli strain EC100 containing the arabinose-inducible expression plasmids pPirAB (PirAB), pPirA (PirA), and pPirB (PirB) from P. asymbiotica and the pBAD30 vector (10) alone were grown at 30°C with aeration to an optical density at 600 nm of 0.4 (11). Protein expression of each clone was induced by adding l-arabinose to a final concentration of 0.2% (wt/vol). The cultures were grown overnight, spun, and washed twice with sterile phosphate-buffered saline and resuspended in an equal volume of phosphate-buffered saline (11). Protein concentrations were measured at the optical density at 600 nm.
All experiments were carried out with first-stage larvae. For the screening tests, 10 first-stage larvae were placed in 5 ml of dechlorinated tap water (Table 1). For the standard oral biological assay, 20 larvae were placed in beakers containing 1,000 ml of dechlorinated tap water (pH 7.0) at 25°C and 75% relative humidity (triplicate experiments). Preliminary tests were conducted with PirAB proteins in two replicates at nine concentrations (Table 2), including the negative control (pBAD30 alone).
TABLE 1.
Bioassay of PirAB from Photorhabdus asymbiotica with Aedes aegypti, Aedes albopictus, and Mesocyclops thermocyclopoides
| P. asymbiotica clone protein or vector | % Mortalitya
|
|||||
|---|---|---|---|---|---|---|
|
A. albopictus
|
A. aegypti
|
M. thermocyclopoides
|
||||
| Day 1 | Day 2 | Day 1 | Day 2 | Day 1 | Day 2 | |
| PirAB | 90 | 100 | 100 | 100 | 0 | 0 |
| pPirABpro | 70 | 80 | 100 | 100 | 0 | 0 |
| pBAD30 negative control vector | 0 | 0 | 0 | 0 | 0 | 0 |
Ten larvae per well.
TABLE 2.
Mortality rate of Aedes aegypti larvae with different PirAB concentrations
| Clone protein or vector concn (106 cells/ml) | % Mortality on day 1
|
|
|---|---|---|
| PirAB clone | pBAD30 negative control vector | |
| 1.3 | 100 | 0 |
| 0.66 | 100 | 0 |
| 0.44 | 100 | 0 |
| 0.33 | 100 | 0 |
| 0.22 | 100 | 0 |
| 0.13 | 100 | 0 |
| 0.066 | 78 | 0 |
| 0.033 | 45 | 0 |
| 0.017 | 18 | 0 |
PirAB from P. luminescens and P. asymbiotica revealed the strongest toxicity against G. mellonella (11). Hence, we have tested the insecticidal toxicity of PirAB proteins from P. asymbiotica expressed in E. coli with the vectors of dengue fever, A. aegypti and A. albopictus. Two distinct expression constructs were tested: pPirAB, in which the PirAB genes are cloned downstream of the arabinose-inducible promoter of pBAD30; and pPirABpro, which also includes the native Pir promoter (11). The recombinant E. coli strain expressing PirAB provides an excellent oral delivery system as these strains can be used directly as the mosquito larva food source. The first-stage larvae of A. aegypti and A. albopictus were killed by PirAB protein (Table 1). A mortality rate of 100% was obtained in both A. albopictus and A. aegypti at day 2, while a 90% mortality rate of A. albopictus mosquitoes was found on day 1 for E. coli pPirAB. In contrast, in E. coli pPirABpro, the toxicity effect is reduced (Table 1). This result shows a trend similar to that found in the work of Waterfield et al. (11) when these strains were injected into the G. mellonella larvae and likely results from a direct repression of PirAB transcription by the native Pir promoter sequence, even when the pBAD arabinose promoter is induced. Furthermore, the bioassay of PirAB protein was performed on M. thermocyclopoides, a common biological control agent of the first-stage larvae of Aedes mosquitoes. A mortality rate of 0% was obtained when testing against the Mesocyclops species. This indicates specificity of PirAB proteins against the dengue vectors. The negative control plasmid (pBAD30) also yielded 0% mortality of the mosquito larvae.
We have selected A. aegypti, the major vector of dengue fever, as a model mosquito to find the optimal concentration of the recombinant E. coli strain expressing PirAB. The lowest concentration (oral biological assay) of PirAB that can kill most A. aegypti larvae (100% mortality on day 1) is 0.33 × 106 cells/ml (Table 2). The negative control showed a 0% mortality rate. This concentration was chosen for further experiments to evaluate Pir proteins from different clones to find the effective titer that could cause death of the mosquito larvae.
The toxicity of PirAB was compared with that of individually expressed PirA or PirB and a mixture of PirA and PirB (designated PirA+PirB, representing a combination of each clone from separate inductions) (Table 3). PirAB shows the strongest larvicidal toxicity with A. aegypti larvae when the larvicidal activities of various types of Pir proteins were compared. The mortality rate was increased and tended to be stable after day 2 with PirB and PirA+PirB proteins. While with PirA, the mortality rate of the larvae was stable after day 4. An increase in the mortality rate with other proteins was still low compared to the toxicity of PirAB by oral bioassay. The mixture of individual PirA and PirB proteins did not reconstitute full activity against mosquito larvae. This finding is in contrast to the work of Waterfield et al. (11) when testing PirA+PirB toxicity for G. mellonella. The full activity against this moth was obtained by injection of PirA+PirB into the insect hemocele. Therefore, PirA, PirB, and PirAB act differently either against different host insects or depending on the delivery route (i.e., injection versus ingestion).
TABLE 3.
Mortality rate of Aedes aegypti larvae with different Pir clones
| Clone protein or vector concn (0.13 × 106 cells/ml) | % Mortality on:
|
||||||
|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | |
| PirAB | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| PirA | 23 | 33 | 35 | 35 | 38 | 38 | 38 |
| PirB | 23 | 30 | 30 | 30 | 30 | 30 | 30 |
| PirA+PirB | 15 | 20 | 20 | 20 | 20 | 20 | 20 |
| pBAD30 negative control vector | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
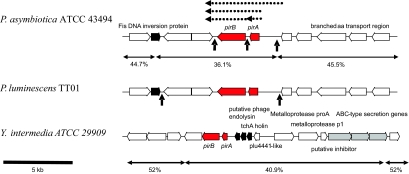
The predicted amino acid sequence of PirA shows 86% identity over 138 amino acids to P. luminescens subsp. laumondii TT01 PirA. PirB also shows significant BLAST X matches to P. luminescens, with 94% identity over 419 amino acids. The interesting point is that both PirA and PirB alone express a low level of toxicity compared to PirAB protein. Even with the expression of the combination of both clones (PirA+PirB), the mortality rate of A. aegypti was still low in comparison with the bioassay with PirAB. Parts of the DNA sequences that are missing from PirA+PirB DNA sequences (individual clone) but included in PirAB were run against NCBI database. The DNA sequences of P. asymbiotica between PirA and PirB contain an enterobacterial repeat intergenic consensus (ERIC) sequence, examples of which are found across the genomes of all Enterobacteriaceae. ERIC sequences have recently been implicated in mRNA stability and expression (5). We suggest that this ERIC element may be important for the strong expression of PirA and PirB and subsequent toxicity against A. aegypti larvae, although this needs further study. Interestingly, comparisons of PirAB with the NCBI database have shown that close homologues are also present in the genome of Yersinia intermedia ATCC 29909 (YintA_01001147 and YintA_0100114788) and are seen in cDNA clones from the beetle Leptinotarsa, suggesting that this toxin family may be more widespread than initially suspected. The genomic loci of PirAB genes in the two Photorhabdus strains (with the positions of the ERIC elements) and also that of Y. intermedia are shown in Fig. 1. These regions are GC skewed, suggesting horizontal movement. The presence of phage genes in the Yersinia Pir locus and of a Fis-type DNA recombinase adjacent to the Photorhabdus operons further supports the suggestion that these genes have been acquired by horizontal transmission. It is possible that the ERIC elements also played a role in acquisition by recA-dependent recombination in Photorhabdus. In conclusion, our preliminary results suggest that the PirAB proteins may serve as biopesticides for controlling the dengue vector. Further study is needed to evaluate the biosafety properties of these proteins before they can be recommended as biological control agents.
FIG. 1.
Comparison of the genomic organization of the pirAB operons in two species of Photorhabdus and one strain of Yersinia. PirAB genes are filled in red. Note that the presence of mobile element-related sequences (filled in black) and the G+C content of the regions suggest recent horizontal acquisition of these genes. Also, note that the Yersinia pirAB genes are tightly linked to a type I secreted metalloprotease operon, suggesting a larger pathogenicity island. ERIC sequences in the Photorhabdus operons are indicated by vertical arrows. Dotted horizontal arrows show the PCR-generated clones tested in this work.
Acknowledgments
We thank Kitti Thienthong for rearing the mosquito colony and Natchaya Klinpikul for rearing the copepod colony used in this study.
This work was supported by Mahidol University Research grant SCBI-2547-27.
Footnotes
Published ahead of print on 8 May 2009.
REFERENCES
- 1.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 2.Boemare, N. E., A. Givaudan, M. Brehelin, and C. Laumond. 1997. Symbiosis and pathogenicity of nematode-bacterium complexes. Symbiosis 22:21-45. [Google Scholar]
- 3.Chattopadhyay, A., N. B. Bhatnagar, and R. Bhatnagar. 2004. Bacterial insecticidal toxins. Crit. Rev. Microbiol. 30:33-54. [DOI] [PubMed] [Google Scholar]
- 4.Daborn, P. J., N. R. Waterfield, C. P. Silva, C. P. Au, S. Sharma, and R. H. Ffrench-Constant. 2002. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99:10742-10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Gregorio, E., G. Silvestro, M. Petrillo, M. S. Carlomagno, and P. P. Di Nocera. 2005. Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: genomic organization and functional properties. J. Bacteriol. 187:7945-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duchaud, E. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 7.Fischer-Le, S. M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. aksurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. P. temperata subsp. nov., and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 8.Gerrard, J. G., S. A. Joyce, D. J. Clarke, R. H. ffrench-Constant, G. R. Nimmo, D. F. Looke, E. J. Feil, L. Pearce, and N. R. Waterfield. 2006. Nematode symbiont for Photorhabdus asymbiotica. Emerg. Infect. Dis. 12:1562-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubler, D. J. 1989. Dengue, p. 223-260. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology, vol. 2. CRC Press, Boca Raton, FL. [Google Scholar]
- 10.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterfield, N. R., S. G. Kamita, B. D. Hammock, and R. ffrench-Constant. 2005. The Photorhabdus Pir toxins are similar to a developmentally regulated insect protein but show no juvenile hormone esterase activity. FEMS Microbiol. Lett. 245:47-52. [DOI] [PubMed] [Google Scholar]
- 12.Weissfeld, A. S., R. J. Halliday, D. E. Simmons, E. A. Trevino, P. H. Vance, C. M. O'Hare, E. G. Sawers, R. Kern, R. D. Koy, K. Hodde, M. Bing, C. Lo, J. Gerrard, R. Vohra, and J. Harper. 2005. Photorhabdus asymbiotica, a pathogen emerging on two continents that proves that there is no substitute for a well-trained clinical microbiologist. J. Clin. Microbiol. 43:4152-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]