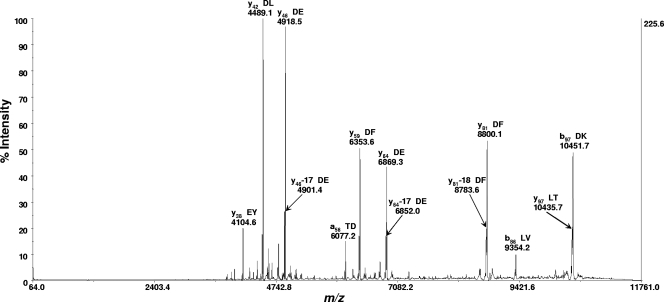
FIG. 3.
MS-MS spectrum (as displayed in commercial instrument software) of the protein biomarker ion at m/z 11138.9 (see Fig. 2) analyzed with the MALDI-TOF-TOF MS instrument (in reflectron mode) using sinapinic acid as the MALDI matrix. Fragmentation was by postsource dissociation. The precursor ion suppressor was “on.” This protein biomarker had been previously identified by bottom-up proteomics as thioredoxin (12). Prominent fragment ions are identified by their m/z, ion type and number, and amino acid residues adjacent to the site of polypeptide backbone cleavage resulting in the fragment ion. Many of the fragment ions are the result of polypeptide cleavage adjacent to an aspartic acid or glutamic acid residue. The MS-MS data were centroided before they were exported as an ASCII-formatted file for upload into the USDA software database.