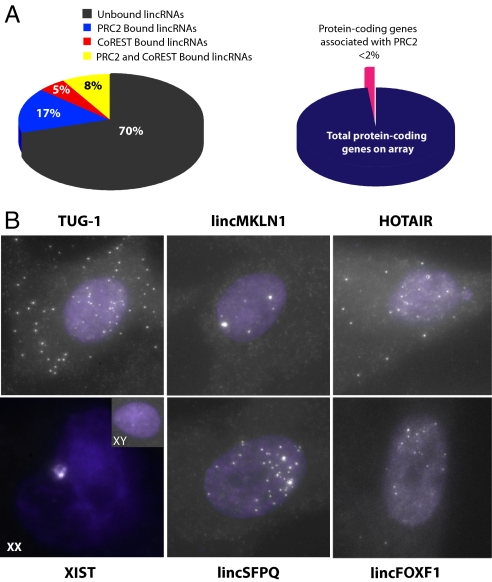
Fig. 3.
Diversity and nuclear localization of chromatin associated lincRNAs. (A) Many lincRNAs, but not protein-coding mRNAs are physically associated with chromatin-modifying complexes. (Left) Pie chart representing the percentage of unique lincRNAs expressed in all 3 tested cell types (hFF, hLF, and HeLa) that are bound only to PRC2 (blue), only to CoREST (red), bound by both PRC2 and CoREST (yellow), and those not bound by either complex (black). (Right) Pie chart indicating the percentage of protein-coding genes (pink) reproducibly bound to PRC2 and or CoREST in all 3 cell types relative to the total number of expressed protein-coding genes (dark blue). (B) Subcellular localization analysis of lincRNAs by RNA FISH demonstrates localization of lincRNAs to the nucleus. Each panel represents the in situ hybridization of ≈40 fluorescently labeled DNA oligos with complementarity to the interrogated lincRNA. RNA FISH experiments were performed in male hFF for each represented lincRNA (XIST, HOTAIR, TUG-1, lincMKLN-1, lincFOXF1, and lincSFPQ), and also in female hLF for XIST (XX). White “speckles” indicate the subcellular localization of each lincRNA. The nuclear compartment is demarked by DAPI staining (purple).