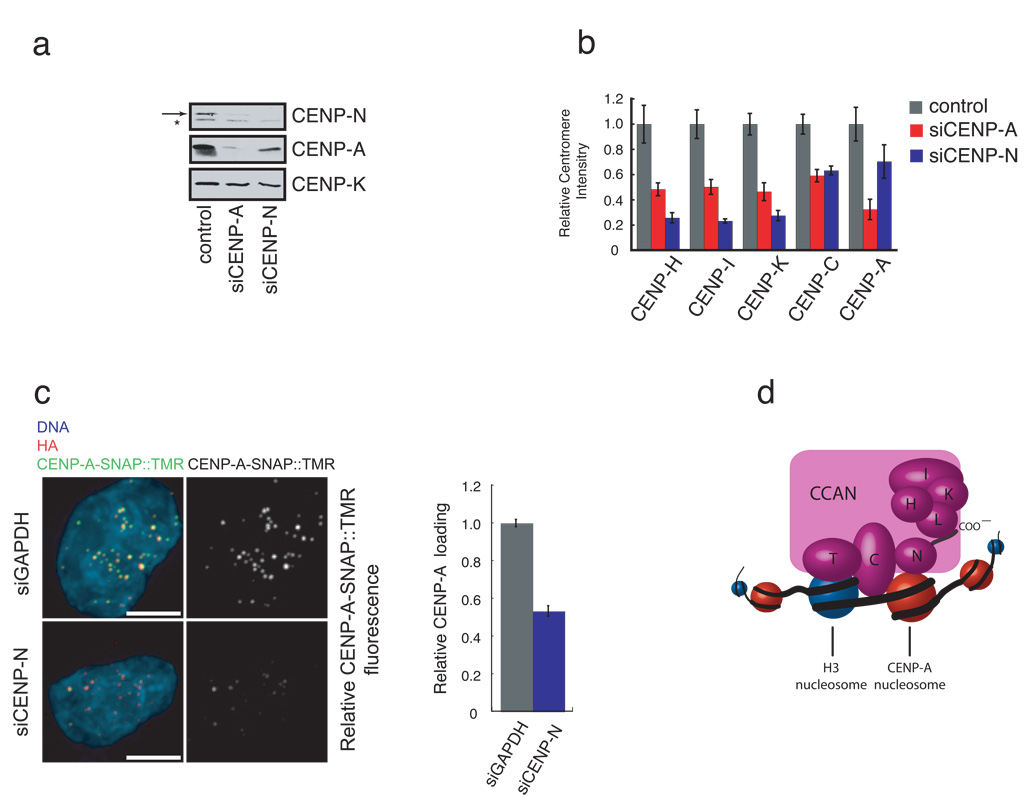
Figure 4. Depletion of CENP-N affects centromere assembly.
a) CENP-N depletion leads to a reduction in CENP-A levels. HeLa cells were treated with the indicated siRNA for 56 hours and the abundance of the indicated centromere protein was determined by western blotting. Arrow (top panel) indicates the position of CENP-N. A background band (*) was present in the anti-CENP-N western blot. CENP-K is included as a loading control. b) CENP-N depletion causes centromere assembly defects. HeLa cells were treated as in (a), except that the centromere fluorescence intensity of the indicated protein was measured. Error bars represent SEM for three independent experiments including >150 centromeres from >10 cells for each experiment. c) Reduced CENP-A assembly after CENP-N RNAi. CENP-A-SNAP cells were synchronized, transfected with siRNA’s against CENP-N or GAPDH and assayed for CENP-A loading by specifically labeling nascent CENP-A-SNAP using TMR-Star as outlined in Figure S5c. Representative images are shown (left panel). HA labels steady state pool of CENP-A-SNAP and is used as centromere marker. Scale bar is 5 µm. Fluorescence intensity of 200 centromeres from 20 cells was quantified in 3 experiments and normalized to the GAPDH siRNA control. Error bars represent SEM. d) A model depicting the multiple roles of CENP-N in CENP-A nucleosome recognition, centromere assembly through the recruitment of CCAN proteins via the CENP-N carboxy-terminal region, and propagation of centromeric chromatin through the CCAN dependent regulation of CENP-A nucleosome assembly.