Abstract
Transcription factors with an APETELA2 (AP2) domain have been implicated in various cellular processes involved in plant development and stress responses. Of the 139 AP2 genes predicted in rice (Oryza sativa), we identified 42 genes in our current study that are induced by one or more stress conditions, including drought, high salinity, low temperature, and abscisic acid. Phylogenic analysis of these 42 stress-inducible AP2 genes revealed the presence of six subgroups (I–VI) with distinct signature motifs. Two genes, AP37 and AP59, representing subgroups I and II, respectively, were functionally characterized. Both genes were found to be induced upon 2 h of exposure to drought and high-salinity conditions but to differ in their expression profile upon exposure to low temperature and abscisic acid. The overexpression of AP37 and AP59 in rice under the control of the constitutive promoter OsCc1 increased the tolerance to drought and high salinity at the vegetative stage. Increased tolerance to low temperatures was observed only in OsCc1:AP37 plants. More importantly, the OsCc1:AP37 plants showed significantly enhanced drought tolerance in the field, which increased grain yield by 16% to 57% over controls under severe drought conditions, yet exhibited no significant difference under normal growth conditions. In contrast, grain yield in OsCc1:AP59 plants in the field was reduced by 23% to 43% compared with controls under both normal and drought stress conditions. Microarray experiments identified 10 and 38 genes that are up-regulated by AP37 and AP59, respectively, in addition to 37 genes that are commonly induced by both factors. Our results suggest that the AP37 gene has the potential to improve drought tolerance in rice without causing undesirable growth phenotypes.
Drought stress is among the most serious challenges to crop production worldwide. Upon exposure of plants to drought conditions, many stress-related genes are induced, and their products are thought to function as cellular protectants of stress-induced damage (Thomashow, 1999; Shinozaki et al., 2003). The expression of stress-related genes is largely regulated by specific transcription factors. Members of the APETELA2 (AP2), bZIP, zinc finger, and MYB families have been shown to have regulatory roles in stress responses. The rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) genomes code for more than 1,300 transcriptional regulators, accounting for about 6% of the estimated total number of genes in both cases. About 45% of these transcription factors were reported to be from plant-specific families (Riechmann et al., 2000; Kikuchi et al., 2003). One example of such a plant-specific family of transcription factors is APETALA2 (AP2), whose members share a highly conserved DNA-binding domain known as AP2 (Weigel, 1995). AP2 factors appear to be widespread in plants, with the genomes of rice and Arabidopsis predicted to contain 139 and 122 AP2 genes, respectively (Nakano et al., 2006). Members of the AP2 family have been implicated in diverse functions in cellular processes involving flower development, spikelet meristem determinacy, plant growth, and stress tolerance (Chuck et al., 1998; Liu et al., 1998; Haake et al., 2002; Dubouzet et al., 2003; Gutterson and Reuber, 2004). Of these diverse functions, the involvement of the AP2 family in stress response has been relatively well characterized. In particular, CBF/DREB genes from Arabidopsis have been shown to play crucial roles in response to low temperature, salt, and drought stresses in transgenic Arabidopsis (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Jaglo et al., 2001). CBF/DREBs are members of the AP2 family and identifiable by the presence of CBF/DREB signature motifs (PKK/RPAGRxKFxETRHP and DSAWR) directly flanking the AP2 domain (Jaglo et al., 2001). Overexpression of CBF/DREBs in transgenic Arabidopsis increases the transcript levels of stress-related genes and enhances tolerance to drought, high-salinity, and freezing stresses (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Haake et al., 2002). CBF/DREBs are also heterologously effective in canola (Brassica napus; Jaglo et al., 2001), tomato (Solanum lycopersicum; Hsieh et al., 2002), tobacco (Nicotiana tabacum; Kasuga et al., 2004), and rice (Oh et al., 2005b), enhancing stress tolerance in the corresponding transgenic plants. CBF/DREB orthologs have also been identified in canola, tomato, wheat (Triticum aestivum), rye (Secale cereale), barley (Hordeum vulgare), and rice, and all of them are inducible by low-temperature treatments (Jaglo et al., 2001; Choi et al., 2002; Xue, 2002, 2003; Dubouzet et al., 2003; Skinner et al., 2005; Oh et al., 2007). The AP2 gene family from other plant species, including DBF1 and DBF2 (Kizis and Pages, 2002) from maize (Zea mays), AhDREB1 (Shen et al., 2003) from Atriplex hortensis, OPBP1 (Guo et al., 2004) from tobacco, CaPF1 (Yi et al., 2004) from pepper (Capsicum annuum), HvRAF (Jung et al., 2006) from barley, and SodERF3 (Trujillo et al., 2008) from sugarcane (Saccharum officinarum), have been found to be involved in responses to various abiotic stress conditions.
Approximately 20% of rice-growing areas worldwide are prone to drought. Although drought conditions can alter the growth and development of rice at any time during its life cycle, drought stress during reproductive growth directly results in a loss of grain yield. To evaluate improvements in grain yield under drought conditions, it is important to subject the plants to the stress during the transition to the reproductive phase. To date, a number of studies have suggested that overexpression of stress-related genes could improve drought tolerance in rice to some extent (Xu et al., 1996; Garg et al., 2002; Jang et al., 2003; Ito et al., 2006; Hu et al., 2006, 2008; Nakashima et al., 2007). Despite such efforts to develop drought-tolerant rice plants, very few of these have been shown to improve grain yields under field conditions. Examples of positive effects include transgenic rice plants expressing SNAC1 (Hu et al., 2006) and OsLEA3 (Xiao et al., 2007), which was shown to improve grain yield under field drought conditions.
In this study, we identified 42 rice genes encoding transcription factors with the AP2 domain that were stress inducible. Two closely related yet distinct genes, AP37 and AP59, were functionally characterized. The overexpression of these genes in transgenic rice improved the plant tolerance to both drought and high salinity at the vegetative stage. However, increased tolerance to low temperature was observed only in plants overexpressing AP37. These AP37 overexpressors showed significantly enhanced drought tolerance in the field, increasing grain yield by 16% to 57% over the controls under severe drought conditions, yet they displayed no significant difference in yield under normal growth conditions. In contrast, grain yield in OsCc1:AP59 plants was reduced by 23% to 43% under both normal and drought stress conditions. We discuss the similarities and differences between the functions of AP37 and AP59 with respect to stress tolerance and grain yield.
RESULTS
Identification of Stress-Inducible AP2 Transcription Factors in Rice
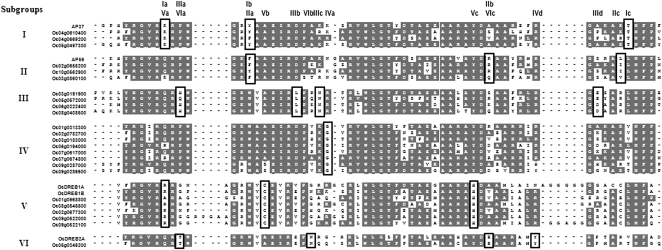
Previously, the rice genome was predicted to contain 139 AP2 domain genes (Nakano et al., 2006). To identify stress-inducible AP2 genes, we performed expression profiling with the Rice 3′-Tiling microarray (GreenGene Biotech) using RNAs from 14-d-old leaves of rice seedlings subjected to drought, high salinity, abscisic acid (ABA), and low temperature. When three replicates were averaged and compared with untreated leaves, a total of 42 genes were found to be up-regulated by 1.6-fold or greater (P < 0.05) by one or more of these stress conditions (Table I). Phylogenic analysis of the amino acid sequences of 42 factors revealed the presence of six subgroups (I–VI), with AP37 assigned to subgroup I, AP59 to subgroup II, OsDREB1A to subgroup V, and OsDREB2A to subgroup VI (Fig. 1; Dubouzet et al., 2003). Thirteen of 42 factors are not classified into any of the six subgroups (Table I; Supplemental Fig. S1). Comparison of the amino acid sequence spanning the AP2 domain identified signature motifs by which these subgroups can be distinguished (Fig. 1). For example, signature motifs Ia, Ic, IIb, and IIc are specific to subgroups I and II, respectively, and motif Ib (IIa) is common to both. In addition to sequence similarity, members of each subgroup are closely related in terms of their response to stress. For example, expression of the genes in subgroups I, II, and V is not induced by ABA, whereas that of the members of subgroups III and VI is not induced by ABA or low temperature.
Table I.
Rice AP2 transcription factor genes up-regulated under stress conditions
Numbers in boldface indicate up-regulation by more than 1.6-fold (P < 0.05) in plants grown under stress conditions.
| Subgroups | Sequence Identifiera | Drought
|
High Salinity
|
ABA
|
Low Temperature
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Meanb | Pc | Meanb | Pc | Meanb | Pc | Meanb | Pc | ||
| I | AP37 | 3.8 | 0.00 | 3.4 | 0.00 | 1.1 | 0.72 | 1.9 | 0.10 |
| Os04g0610400 | 2.1 | 0.01 | 1.5 | 0.17 | −1.2 | 0.65 | 2.9 | 0.02 | |
| Os04g0669200 | 2.8 | 0.01 | 2.6 | 0.02 | 1.3 | 0.59 | 2.9 | 0.04 | |
| Os05g0497300 | 2.7 | 0.00 | 1.8 | 0.05 | 1.1 | 0.86 | 2.5 | 0.04 | |
| II | AP59 | 7.7 | 0.00 | 6.1 | 0.00 | 1.3 | 0.43 | 8.6 | 0.00 |
| Os02g0655200 | 1.4 | 0.27 | 2.8 | 0.01 | 1.2 | 0.63 | −1.1 | 0.91 | |
| Os10g0562900 | 1.3 | 0.27 | 1.1 | 0.92 | 1.3 | 0.52 | 10.1 | 0.00 | |
| Os03g0860100 | 3.6 | 0.00 | 2.4 | 0.02 | −1.4 | 0.39 | 1.2 | 0.72 | |
| III | Os03g0191900 | 12.7 | 0.00 | 4.9 | 0.00 | −1.3 | 0.42 | 1.2 | 0.56 |
| Os05g0572000 | 3.5 | 0.00 | 3.2 | 0.00 | 1.4 | 0.29 | −1.1 | 0.87 | |
| Os06g0222400 | 1.8 | 0.03 | 1.2 | 0.58 | −1.6 | 0.13 | −1.3 | 0.43 | |
| Os08g0408500 | 9.1 | 0.00 | 5.2 | 0.00 | 1.0 | 0.98 | 1.2 | 0.61 | |
| IV | Os01g0313300 | 1.5 | 0.29 | 1.3 | 0.69 | 3.2 | 0.02 | 4.2 | 0.03 |
| Os02g0782700 | 1.2 | 0.41 | 1.5 | 0.14 | 1.9 | 0.04 | −1.0 | 0.93 | |
| Os03g0183000 | 2.0 | 0.01 | 2.6 | 0.00 | 12.8 | 0.00 | 1.4 | 0.30 | |
| Os06g0194000 | 1.6 | 0.04 | 1.7 | 0.04 | 1.6 | 0.17 | −1.3 | 0.51 | |
| Os07g0617000 | 2.3 | 0.01 | 2.7 | 0.01 | 4.0 | 0.00 | 2.1 | 0.07 | |
| Os07g0674800 | 1.3 | 0.33 | 1.8 | 0.03 | −1.0 | 0.96 | 1.8 | 0.09 | |
| Os09g0287000 | −1.9 | 0.19 | −1.2 | 0.89 | 22.0 | 0.00 | 2.0 | 0.30 | |
| Os09g0286600 | 1.2 | 0.55 | −1.0 | 0.99 | 2.3 | 0.02 | 1.2 | 0.59 | |
| V | OsDREB1A | 2.4 | 0.01 | −1.1 | 0.91 | −1.8 | 0.07 | 7.7 | 0.00 |
| OsDREB1B | 9.5 | 0.00 | 6.0 | 0.00 | 1.2 | 0.67 | 3.1 | 0.04 | |
| Os01g0968800 | 6.3 | 0.00 | 5.7 | 0.00 | −1.3 | 0.62 | 7.9 | 0.00 | |
| Os08g0545500 | 2.2 | 0.01 | 1.5 | 0.24 | −1.0 | 0.99 | 1.5 | 0.33 | |
| Os02g0677300 | 1.6 | 0.19 | 1.1 | 0.95 | 1.0 | 0.98 | 8.2 | 0.00 | |
| Os09g0522000 | −1.2 | 0.40 | −1.9 | 0.02 | −3.2 | 0.00 | 51.4 | 0.00 | |
| Os09g0522100 | 1.6 | 0.17 | −1.1 | 0.93 | 1.1 | 0.90 | 6.4 | 0.01 | |
| VI | OsDREB2A | 3.0 | 0.01 | 3.8 | 0.01 | 2.2 | 0.07 | −1.6 | 0.41 |
| Os05g0346200 | 2.9 | 0.00 | 2.2 | 0.00 | 1.5 | 0.09 | 1.2 | 0.50 | |
| Others | Os06g0166400 | 2.8 | 0.00 | 2.5 | 0.01 | 1.8 | 0.05 | 1.1 | 0.83 |
| Os02g0797100 | 3.3 | 0.01 | 1.5 | 0.37 | −1.0 | 0.95 | 1.5 | 0.48 | |
| Os01g0131600 | 1.8 | 0.01 | 1.2 | 0.53 | 1.1 | 0.70 | 1.0 | 0.99 | |
| Os09g0309700 | 1.2 | 0.43 | 1.5 | 0.12 | 2.1 | 0.01 | −1.0 | 0.98 | |
| Os01g0868000 | 2.6 | 0.02 | 1.8 | 0.16 | 1.6 | 0.30 | 1.7 | 0.32 | |
| Os02g0546600 | 9.7 | 0.01 | 3.9 | 0.07 | 1.4 | 0.75 | 2.3 | 0.38 | |
| Os04g0398000 | 2.0 | 0.01 | 2.3 | 0.01 | 1.5 | 0.17 | −1.0 | 0.99 | |
| Os12g0168100 | 5.1 | 0.00 | 3.9 | 0.00 | 1.7 | 0.26 | 1.3 | 0.67 | |
| Os02g0764700 | 9.4 | 0.00 | 4.2 | 0.00 | −1.0 | 0.96 | 6.9 | 0.01 | |
| Os08g0474000 | 21.2 | 0.00 | 14.1 | 0.00 | 2.2 | 0.05 | 26.7 | 0.00 | |
| Os09g0457900 | 15.2 | 0.00 | 7.7 | 0.00 | 1.2 | 0.67 | 62.5 | 0.00 | |
| Os08g0537900 | 3.1 | 0.00 | 3.2 | 0.01 | 5.1 | 0.00 | −1.2 | 0.65 | |
| Os08g0521600 | 4.7 | 0.00 | 3.0 | 0.01 | −1.1 | 0.85 | −1.1 | 0.84 | |
Numbers for full-length cDNA sequences of the corresponding genes.
Numbers represent means of three independent biological replicates. These microarray data sets can be found at http://www.ncbi.nlm.nih.gov/geo/ (Gene Expression Omnibus).
P values were analyzed by one-way ANOVA.
Figure 1.
Alignment of AP2 domain sequences from 29 stress-inducible rice genes. Deduced amino acid sequences of the AP2 domains of the 29 genes listed in Table I were aligned using the ClustalW program. Identical and conserved residues are highlighted in gray. Signature motifs are indicated by boxes identified on the top as follows: Ia to Ic, IIa to IIc, IIIa to IIId, IVa, Va to Vc, and VIa to VId for subgroups I to VI, respectively.
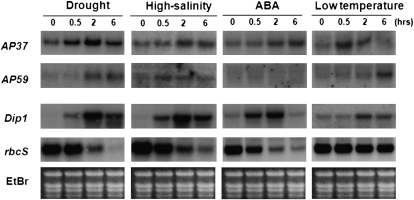
Two genes, AP37 (AK061380) and AP59 (AK073812), representing subgroups I and II, respectively, were functionally characterized in this study. The transcript levels of AP37 and AP59 were measured by RNA gel-blot analysis using total RNAs from leaf tissues of 14-d-old seedlings exposed to high salinity, drought, ABA, and low temperature (Fig. 2). The expression of both AP37 and AP59 was found to be induced after 2 h of exposure to high-salinity and drought stress. AP37 differs from AP59 in its response to low temperature and ABA. The expression of the former responded rapidly to low temperature and was induced by ABA, whereas the latter responded slowly to low temperature and was not induced by ABA. This is somewhat inconsistent with the microarray results, which indicated that AP37 is not induced by ABA. This discrepancy may be due to variation in the stress treatments. Overall, our results showed that AP37 and AP59 are stress-inducible AP2 genes that are closely related yet different in their expression profiles.
Figure 2.
Expression of AP37 and AP59 in response to stress conditions in rice. Ten micrograms of total RNA was prepared from leaf tissues of 14-d-old seedlings exposed to drought, high salinity, ABA, or low temperature for the indicated time points: for drought stress, the seedlings were air dried at 28°C; for high-salinity stress, seedlings were exposed to 400 mm NaCl at 28°C; for low-temperature stress, seedlings were exposed to 4°C; for ABA stress, seedlings were exposed to a solution containing 100 μm ABA. Total RNAs were blotted and hybridized with the AP37 and AP59 gene-specific probes. The blots were then reprobed for the Dip1 (Oh et al., 2005b) and rbcS (Jang et al., 1999) genes, which were used as markers for up- and down-regulation, respectively, of key genes following stress treatments. Ethidium bromide (EtBr) staining was used to determine equal loading of RNAs.
Stress Tolerance of OsCc1:AP37 and OsCc1:AP59 Plants at the Vegetative Stage
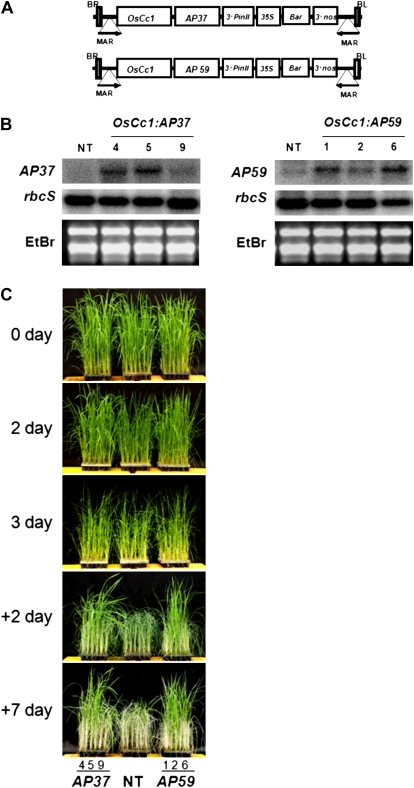
To enable the overexpression of the AP37 and AP59 genes in rice, their full-length cDNAs were isolated and linked to the OsCc1 promoter for constitutive expression (Jang et al., 2002), generating the constructs OsCc1:AP37 and OsCc1:AP59 (Fig. 3A). These constructs were then introduced into rice by Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994), which yielded 15 to 20 independent transgenic lines per construct. Transgenic T1-4 seeds were collected, and three independent T4-5 homozygous lines of both OsCc1:AP37 and OsCc1:AP59 plants were selected for further analysis. All of the transgenic lines grew normally with no stunting. The transcript levels of AP37 and AP59 in the OsCc1:AP37 and OsCc1:AP59 plants were determined by RNA gel-blot analysis. For this purpose, total RNAs were extracted from leaf tissues of 14-d-old seedlings grown under normal growth conditions (Fig. 3B). Transcript levels of AP37 and AP59 were clearly enhanced at various levels in different transgenic lines as compared with those in the nontransgenic (NT) controls. To investigate whether the overexpression of AP37 and AP59 correlated with stress tolerance in rice, 4-week-old transgenic plants and NT controls were exposed to drought stress (Fig. 3C). The NT plants started to show visual symptoms of drought-induced damage, such as leaf rolling and wilting with a concomitant loss of chlorophylls, at an earlier stage than the OsCc1:AP37 and OsCc1:AP59 plants. The transgenic plants also recovered faster than the NT plants upon rewatering. Consequently, the NT plants remained severely affected by the time at which all of the transgenic lines had fully recovered except for the OsCc1:AP37-9 line. This line recovered slower than the other transgenic lines, probably due to a lower level of transgene expression than in the others (Fig. 3, B and C). Levels of transgene expression in the OsCc1:AP59-2 line were also lower than those of others, while its phenotype was comparable to those of the other lines (Fig. 3, B and C). Thus, the difference in transgenic phenotype does not always reflect different levels of mRNA.
Figure 3.
Production of OsCc1:AP37 and OsCc1:AP59 transgenic rice plants. A, The OsCc1:AP37 and OsCc1:AP59 plasmids consist of OsCc1 promoter (Jang et al., 2002), a constitutive promoter linked to the AP37 and AP59 coding region, respectively, the 3′ region of the potato (Solanum tuberosum) proteinase inhibitor II gene (3′PinII), and a gene expression cassette that contains the 35S promoter, the bar coding region, and the 3′ region of the nopaline synthase gene (3′nos). The entire expression cassette is flanked by the 5′ matrix attachment region (MAR) of the chicken lysozyme gene (Oh et al., 2005a). BL, Left border; BR, right border. B, RNA gel-blot analysis was performed using total RNAs from young leaves of three homozygous T4 lines of OsCc1:AP37 and OsCc1:AP59 plants and of NT control plants. The blots were hybridized with the AP37 and AP59 gene-specific probes and reprobed for rbcS. Ethidium bromide (EtBr) staining was used to determine equal loading of RNAs. C, Appearance of transgenic plants during drought stress Three independent homozygous T4 lines of OsCc1:AP37 and OsCc1:AP59 plants and NT controls were grown in a greenhouse for 4 weeks and subjected to drought stress treatments. Four-week-old transgenic and NT plants were subjected to 3 d of drought stress, followed by 2 and 7 d of rewatering in the greenhouse, respectively. Photographs were taken at the indicated time points. + denotes the number of rewatering days under normal growth conditions.
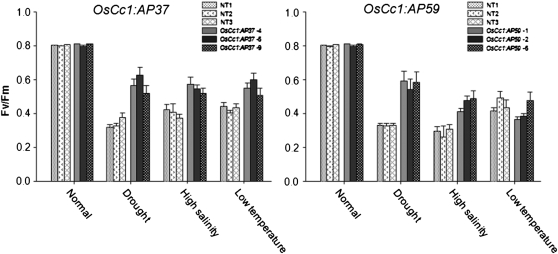
To further verify the stress-tolerance phenotype, we measured the Fv/Fm values of the transgenic and NT control plants, all at the vegetative stage (Fig. 4). The Fv/Fm values represent the maximum photochemical efficiency of PSII in a dark-adapted state, where Fv stands for variable fluorescence and Fm stands for maximum fluorescence. The Fv/Fm levels were about 30% and 20% higher in the OsCc1:AP37 and OsCc1:AP59 plants, respectively, than in the NT plants under drought and high-salinity conditions. Under low-temperature conditions, in contrast, the Fv/Fm levels were 15% higher in the OsCc1:AP37 plants than in the NT plants, whereas the levels in the OsCc1:AP59 plants were at similar levels to those of the NT plants. Hence, our results indicate that the overexpression of AP37 and AP59 in transgenic rice increases the tolerance of these plants to drought and high-salinity stress conditions during the vegetative stage but that an increased tolerance to low temperature occurs only in plants overexpressing AP37.
Figure 4.
Changes in chlorophyll fluorescence (Fv/Fm) of rice plants under drought, high-salinity, and low-temperature stress conditions. Three independent homozygous T4 lines of OsCc1:AP37 and OsCc1:AP59 plants and NT controls grown in MS medium for 14 d were subjected to various stress conditions as described in “Materials and Methods.” After stress treatments, the Fv/Fm values were measured using a pulse modulation fluorometer (mini-PAM; Walz). All plants were grown under continuous light of 150 μmol m−2 s−1 prior to stress treatments. Each data point represents the mean ± se of triplicate determinations (n = 10).
Identification of Genes Up-Regulated by Overexpressed AP37 and AP59
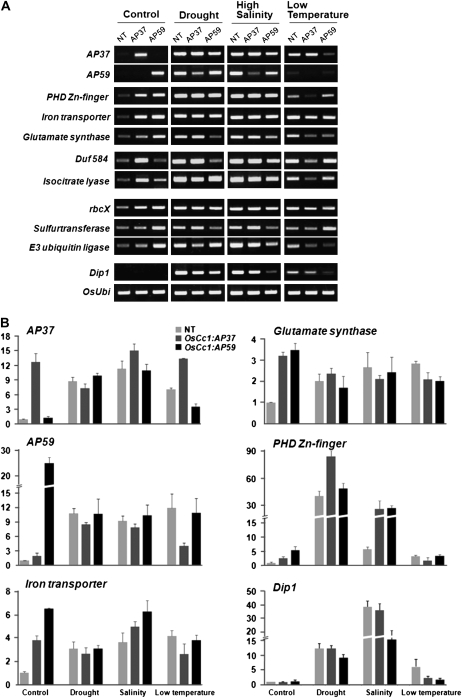
To identify genes that are up-regulated by the overexpression of AP37 and AP59, we performed expression profiling of OsCc1:AP37 and OsCc1:AP59 plants in comparison with NT controls under normal growth conditions. Expression profiling with the Rice 3′-Tiling microarray was conducted using RNA samples extracted from 14-d-old leaves of these transgenic plants and NT controls, all grown under normal growth conditions. Each data set was obtained from three biological replicates. As listed in Supplemental Table S1, statistical analysis of each data set using one-way ANOVA identified 85 genes that are up-regulated by AP37 and/or AP59 with 3-fold or greater induction in the transgenic plants than in NT plants (P < 0.05). Specifically, a total of 37 genes were found to be commonly activated by AP37 and AP59, whereas 10 and 38 genes are specific to AP37 and AP59, respectively. Based on our microarray data shown in Table I and Supplemental Table S1, we selected eight stress-inducible genes out of 37 common target genes and verified their AP37- and AP59-dependent expression patterns under normal growth conditions by reverse transcription (RT)-PCR (Fig. 5A, control). To test whether their expression levels increased further under stress conditions, we measured the transcript levels of the eight genes in OsCc1:AP37, OsCc1:AP59, and NT plants after exposure to drought, high-salinity, and low-temperature conditions (Fig. 5). In NT plants, all of the target genes were induced at various levels within 2 h of the stress treatments. In the transgenic plants, in contrast, the transcript levels of many genes are lower under stress conditions in comparison with stress-treated NT plants. The same is true for the AP59 gene in OsCc1:AP37 plants exposed to drought, high-salinity, and low-temperature conditions and also for AP37 in OsCc1:AP59 plants grown under low-temperature conditions. Such lower levels of target gene transcripts in OsCc1:AP37 and OsCc1:AP59 plants than in NT plants under stress conditions were not unexpected, because the transgenic plants were less affected by stress damage compared with the NT controls. The transgenic plants were more tolerant to stress than the NT controls at the time of stress treatments; hence, stress-induced expression levels of target genes were smaller in the transgenic plants than in NT controls. We repeated the experiments shown in Figure 5A using real-time PCR, obtaining results similar to those of RT-PCR (Fig. 5B; Supplemental Fig. S2). Overall, our results suggest that the AP37 and AP59 genes enhance stress tolerance differently by activating distinct groups of stress-regulated genes.
Figure 5.
Regulated expression of stress-related genes in OsCc1:AP37, OsCc1:AP59, and NT plants under normal and stress conditions. Homozygous T4 lines of OsCc1:AP37, OsCc1:AP59, and NT control rice plants were grown in a greenhouse for 14 d. Transgenic and NT plants were then treated with various stress conditions as described in the legend to Figure 2. A, Transcript levels of AP37, AP59, and eight target genes were determined by RT-PCR (using primers listed in Supplemental Table S3). B, Transcript levels of AP37, AP59, and three target genes were determined by quantitative RT-PCR (using primers listed in Supplemental Table S3). Dip1 (Oh et al., 2005b) was used as a marker for the up-regulation of key genes following stress treatments. The rice ubiquitin gene (OsUbi) was used as an equal loading control.
Overexpression of AP37 Increases Rice Grain Yield under Drought Conditions
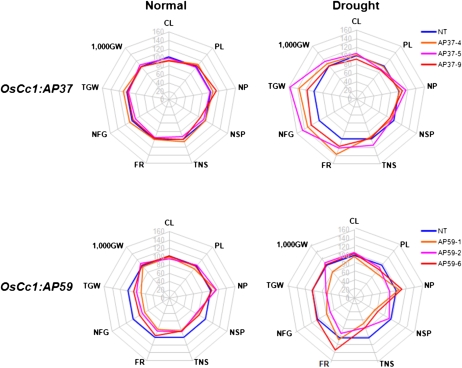
A phenotypic evaluation of OsCc1:AP37, OsCc1:AP59, and NT control plants revealed no major differences in the vegetative growth of the entire plants. To then investigate whether the overexpression of AP37 and AP59 improved the rice grain yield of transgenic plants under field conditions, we transplanted three independent T5 homozygous lines of the OsCc1:AP37 and OsCc1:AP59 plants, together with their respective NT controls, to a paddy field and grew them to maturity. A completely randomized design with two replicates was employed. The subsequent evaluation of the yield parameters of these plants revealed that the grain yield of OsCc1:AP37 plants remained similar to that of the NT controls under normal field conditions (Fig. 6; Table II; Supplemental Table S2). In the OsCc1:AP59 plants under the same field conditions, however, total grain weight was reduced by 23% to 43% compared with the NT controls, which appears to be due to decreases in the number of spikelets. These observations prompted us to examine the yield components of the transgenic rice plants grown in the field under drought conditions. Three independent lines of the OsCc1:AP37 and OsCc1:AP59 plants and NT controls were transplanted to a paddy field with a removable rain-off shelter and exposed to drought stress at the panicle heading stage (from 10 d before heading to 20 d after heading). Statistical analysis of the yield parameters showed that the decrease in grain yield under drought conditions was significantly smaller in the OsCc1:AP37 plants than in the controls. Specifically, in the drought-treated OsCc1:AP37 plants, the filling rate was higher than in the drought-treated NT plants by 17% to 36%, which resulted in an increase in the total grain weight by 16% to 57%, depending on the transgenic line (Fig. 6; Table III). In the drought-treated OsCc1:AP59 plants, in contrast, the total grain weight was reduced by about 30% when compared with the drought-treated NT controls, which was similar to the yields observed for these plants under normal growth conditions. Despite the similar levels of drought tolerance at the vegetative stage of the OsCc1:AP37 and OsCc1:AP59 plants, a sharp difference in grain yield under drought conditions is actually not surprising. This is because, unlike in OsCc1:AP37 plants, spikelet development of OsCc1:AP59 plants was significantly affected by the constitutive overexpression of AP59 under both normal and drought conditions, which resulted in a loss of grain yield. Taken together, these results suggest that the overexpression of AP37 confers drought tolerance in rice at the reproductive stage and improves grain yield significantly.
Figure 6.
Agronomic traits for OsCc1:AP37 and OsCc1:AP59 rice plants grown in the field under normal and stress conditions. Spider plots of agronomic traits of three independent homozygous T5 lines of OsCc1:AP37 and OsCc1:AP59 and corresponding NT controls under normal and drought conditions, respectively, were drawn using Microsoft Excel software. Each data point represents a percentage of the mean values (n = 20) listed in Tables II and III. Mean values from NT controls were set at 100% as a reference. CL, Culm length; PL, panicle length; NP, number of panicles per hill; NSP, number of spikelets per panicle; TNS, total number of spikelets; FR, filling rate; NFG, number of filled grains; TGW, total grain weight; 1,000GW, 1,000 grain weight.
Table II.
Analysis of seed production parameters in OsCc1:AP37 and OsCc1:AP59 plants under normal growth conditions
Each parameter value represents the mean ± sd (n = 20) for OsCc1:AP37 and OsCc1:AP59 and the respective NT control plants. The percentage differences (%Δ) between the values for the OsCc1:AP37 or OsCc1:AP59 plants and for the respective NT controls were calculated. P values were determined according to the lsd test.
| Constructs | Panicle Length | No. of Spikelets | Total No. of Spikelets | Filling Rate | No. of Filled Grain | Total Grain Weight |
|---|---|---|---|---|---|---|
| cm | per panicle | per hill | % | per hill | g | |
| NT | 18.32 ± 1.37 | 96.04 ± 12.74 | 1,209.70 ± 137.55 | 90.80 ± 3.84 | 1,098.45 ± 133.26 | 22.51 ± 3.03 |
| AP37-4 | 19.70 ± 1.25 | 95.86 ± 8.19 | 1,294.55 ± 244.75 | 91.23 ± 2.82 | 1,177.65 ± 202.93 | 24.56 ± 5.28 |
| %Δ | 7.53 | −0.19 | 7.01 | 0.47 | 7.21 | 9.11 |
| P | 0.011 | 0.951 | 0.151 | 0.886 | 0.144 | 0.129 |
| AP37-5 | 18.17 ± 1.18 | 90.76 ± 9.50 | 1,136.95 ± 152.66 | 87.15 ± 3.68 | 992.80 ± 151.39 | 21.60 ± 3.83 |
| %Δ | −0.82 | −5.50 | −6.01 | −4.02 | −9.62 | −4.04 |
| P | 0.776 | 0.086 | 0.218 | 0.665 | 0.053 | 0.500 |
| AP37-9 | 19.10 ± 2.48 | 83.68 ± 7.10 | 1,205.95 ± 186.49 | 88.24 ± 2.86 | 1,066.50 ± 182.78 | 22.31 ± 4.39 |
| %Δ | 4.26 | −12.87 | −0.31 | −2.82 | −2.91 | −0.89 |
| P | 0.144 | 0.000 | 0.949 | 0.075 | 0.553 | 0.884 |
| NT | 18.80 ± 1.19 | 98.90 ± 6.99 | 1,327.10 ± 191.16 | 92.84 ± 3.01 | 1,232.55 ± 185.73 | 25.78 ± 4.39 |
| AP59-1 | 16.95 ± 1.14 | 71.39 ± 11.76 | 996.10 ± 181.07 | 73.90 ± 7.57 | 734.75 ± 156.91 | 14.72 ± 4.34 |
| %Δ | −9.84 | −27.81 | −24.94 | −20.39 | −40.38 | −42.88 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| AP59-2 | 18.60 ± 1.50 | 71.70 ± 5.37 | 1,044.20 ± 241.54 | 78.30 ± 6.08 | 811.20 ± 167.09 | 17.97 ± 4.42 |
| %Δ | −1.06 | −27.50 | −21.31 | −15.65 | −34.18 | −30.27 |
| P | 0.622 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| AP59-6 | 18.45 ± 1.23 | 80.13 ± 10.63 | 1,045.10 ± 186.85 | 88.01 ± 3.60 | 919.25 ± 165.62 | 19.70 ± 4.19 |
| %Δ | −1.87 | −18.97 | −21.24 | −5.19 | −25.41 | −23.58 |
| P | 0.389 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 |
Table III.
Analysis of seed production parameters in OsCc1:AP37 and OsCc1:AP59 plants under drought stress conditions
Each parameter value represents the mean ± sd (n = 20) for OsCc1:AP37 and OsCc1:AP59 and the respective NT control plants. The percentage differences (%Δ) between the values for the OsCc1:AP37 or OsCc1:AP59 plants and for the respective NT controls were calculated. P values were determined according to the lsd test.
| Constructs | Panicle Length | No. of Spikelets | Total No. of Spikelets | Filling Rate | No. of Filled Grain | Total Grain Weight |
|---|---|---|---|---|---|---|
| cm | per panicle | per hill | % | per hill | g | |
| NT | 19.50 ± 2.38 | 98.34 ± 14.52 | 1,526.76 ± 753.88 | 60.15 ± 16.39 | 871.75 ± 349.09 | 15.20 ± 7.29 |
| AP37-4 | 19.10 ± 1.51 | 92.49 ± 11.71 | 1,443.80 ± 662.61 | 82.24 ± 10.53 | 1,144.40 ± 472.48 | 20.60 ± 9.67 |
| %Δ | −2.05 | −5.94 | −5.43 | 36.71 | 31.27 | 35.52 |
| P | 0.703 | 0.458 | 0.819 | 0.020 | 0.261 | 0.273 |
| AP37-5 | 17.50 ± 0.91 | 93.91 ± 6.73 | 1,741.25 ± 212.78 | 72.88 ± 10.67 | 1,262.50 ± 203.76 | 23.87 ± 3.75 |
| %Δ | −10.25 | −4.50 | 14.04 | 21.15 | 44.82 | 57.07 |
| P | 0.087 | 0.592 | 0.576 | 0.171 | 0.134 | 0.105 |
| AP37-9 | 16.85 ± 0.75 | 87.29 ± 11.10 | 1,483.75 ± 106.22 | 70.92 ± 11.66 | 1,059.75 ± 240.49 | 17.70 ± 4.75 |
| %Δ | −13.46 | −11.23 | −2.81 | 17.89 | 21.56 | 16.44 |
| P | 0.030 | 0.194 | 0.910 | 0.242 | 0.455 | 0.624 |
| NT | 19.75 ± 2.06 | 97.01 ± 12.27 | 1,538.25 ± 737.31 | 60.31 ± 16.38 | 895.25 ± 307.00 | 15.45 ± 6.82 |
| AP59-1 | 15.37 ± 2.49 | 54.86 ± 20.71 | 1,001.25 ± 589.52 | 63.20 ± 10.24 | 663.75 ± 481.75 | 9.93 ± 10.41 |
| %Δ | −22.15 | −43.45 | −34.90 | 4.78 | −25.85 | −35.76 |
| P | 0.008 | 0.030 | 0.199 | 0.781 | 0.356 | 0.324 |
| AP59-2 | 18.37 ± 0.94 | 92.25 ± 37.81 | 1,133.75 ± 431.63 | 53.66 ± 15.71 | 579.25 ± 170.12 | 10.40 ± 4.35 |
| %Δ | −6.96 | −4.89 | −26.29 | −11.01 | −35.29 | −32.68 |
| P | 0.331 | 0.785 | 0.325 | 0.526 | 0.215 | 0.366 |
| AP59-6 | 17.17 ± 1.76 | 60.89 ± 12.58 | 1,137.33 ± 280.32 | 78.86 ± 14.19 | 902.33 ± 317.86 | 15.37 ± 7.36 |
| %Δ | −13.08 | −37.23 | −26.06 | 30.76 | 0.79 | −0.53 |
| P | 0.105 | 0.075 | 0.365 | 0.119 | 0.979 | 0.989 |
DISCUSSION
In this study, expression profiling using RNAs from stress-treated rice plants identified 42 AP2 domain factors that are stress inducible (Table I). Alignment of these stress-inducible factors revealed six subgroups within which the members are more closely related, suggesting a common function during the stress response. The overexpression of AP37 (OsCc1:AP37) and AP59 (OsCc1:AP59), representative members of subgroups I and II, respectively, increased the rice plant tolerance to drought and high salinity at the vegetative stage. Increased tolerance to low temperature was observed only in OsCc1:AP37 plants, suggesting a functional difference between the two closely related AP2 factors in the stress response. Given the different numbers of target genes up-regulated in OsCc1:AP37 and OsCc1:AP59 plants, the difference in their response to low temperature was not unexpected. Our microarray experiments identified 10 and 38 putative target genes that are specific to the AP37 and AP59 proteins, respectively, in addition to a further 37 target genes that appeared to be common to both factors (Supplemental Table S1). Similarly contrasting results were previously obtained for two closely related AP2 factors, HvCBF4 from barley and CBF3/DREB1A from Arabidopsis. The overexpression of HvCBF4 and CBF3/DREB1A in rice confers increased tolerance to drought, high salinity, and low temperature, and in the case of low temperature this effect was more pronounced in plants overexpressing HvCBF4 (Oh et al., 2007). The composition of the target rice genes was also different between the HvCBF4 and CBF3/DREB1A plants.
The expression patterns of the AP37 and AP59 genes are similar under drought and high-salinity conditions but different under low-temperature and ABA stress, consistent with the observed differences between these genes in conferring low-temperature tolerance. The AP37 transcript levels were rapidly increased in rice plants within 30 min of exposure to stress conditions, similar to the reported observations for the OsDREB1A transcripts (Dubouzet et al., 2003). The OsDREB1A gene was classified in subgroup V in this analysis, the members of which have a similar expression pattern to the genes in subgroups I and II (Table I). Consistent with our results for OsCc1:AP37 rice plants, the overexpression of OsDREB1A in rice (Ito et al., 2006) and in Arabidopsis (Dubouzet et al., 2003) has been shown previously to confer tolerance to low temperature, in addition to drought and high-salinity resistance. The overexpression of OsDREB1A in rice induced the accumulation of soluble sugars, including raffinose, Suc, Glc, and Fru, which may act as osmoprotectants (Ito et al., 2006). The target genes we identified in OsCc1:AP37 plants included genes that function in carbon metabolism such as phosphogluconate aldolase, phosphoglucomutase, UDP-glucosyl transferase, and isocitrate lyase. These genes may similarly increase the content of soluble sugars. It is also generally accepted that excessive amounts of reactive oxygen species are generated upon exposure of plants to stress stimuli, and these must be removed in order to maintain cellular homeostasis. Overexpression of JERF3, a tomato ortholog of our subgroup IV (Table I), was shown previously to enhance stress tolerance by increasing the expression of genes encoding antioxidant enzymes (Wu et al., 2008). Similarly, the expression levels of several antioxidant genes, such as thioredoxin, peroxidase, and ascorbate oxidoreductase, were found to be increased in our OsCc1:AP37 and OsCc1:AP59 transgenic rice plants, which may indicate the activation of a reactive oxygen species-scavenging system. Loss-of-function mutants on AP37 and AP59 may provide us with further evidence of molecular mechanisms for stress tolerance, although the presence of many homologous AP2 domain genes may not allow knockout phenotypes to be displayed.
Grain yield from rice plants is severely affected when they are exposed to drought stress at the reproductive stage. Therefore, it was important to examine the effects of drought stress on grain yield at this stage of growth in our transgenic plants. It was also important to use transgenic lines that were not genetically segregating under field conditions. It is relatively straightforward to identify segregating families of transgenic rice plants up to the T4 generation in the field, even though they are homozygous for a transgene. To evaluate whether any improvements in grain yield had occurred in our transgenic rice under drought conditions, we transplanted T5 homozygous lines of OsCc1:AP37 and OsCc1:AP59 plants to the field in 2008, which had been prescreened in the field for segregation in 2007. The plants were exposed to drought stress at the panicle heading stage from 10 d prior to heading to 20 d after heading in field conditions. The OsCc1:AP37 plants showed significantly enhanced drought tolerance in the field, with a grain yield of 16% to 57% higher than the controls under severe drought conditions yet displayed no significant differences under normal growth conditions (Fig. 6; Tables II and III). In contrast, grain yield from the OsCc1:AP59 plants in the field was reduced by 23% to 43% compared with the controls under both normal and drought stress conditions. The decrease in grain yield in OsCc1:AP59 plants was primarily due to the disrupted spikelet development caused by augmented AP59 expression. The vegetative growth of both the OsCc1:AP37 and OsCc1:AP59 plants was visually indistinguishable from that of the NT controls. Importantly, the overexpression of AP37 in rice was effective against drought stress at the reproductive stage as well as at the vegetative stage. In addition, the overexpression of AP37 does not seem to affect the development of reproductive organs while conferring stress tolerance in transgenic plants. Development of the panicle and/or spikelet meristem is repressed in rice under drought conditions, resulting in a reduction in the number of panicles and/or spikelets (Boonjung and Fukai, 1996; Wopereis et al., 1996; Asch et al., 2005). The lower decreases in the filling rate and in the number of spikelets of OsCc1:AP37 plants under drought conditions implied that the developmental processes for panicles and spikelets had been protected from drought stress, indicating drought tolerance at the reproductive stage. The overexpression of AP59, in contrast, was ineffective in increasing grain productivity, yet it conferred stress tolerance in rice plants at the vegetative stage. To date, the potential impact of homeotic genes like the AP2 factors upon grain yield have received relatively little attention, because of their negative effects on fertility, plant growth, and development. It is thus important to evaluate agronomic traits in transgenic crops throughout all stages of plant growth to address the advantages of using such homeotic genes for improving stress tolerance. Overall, our results here demonstrate that AP37 can improve rice grain yield under drought conditions without conferring an undesirable growth phenotype.
MATERIALS AND METHODS
Plasmid Construction and Transformation of Rice
The expression plasmids OsCc1:AP37 and OsCc1:AP59 contained the bar gene under the control of the cauliflower mosaic virus 35S promoter to enable herbicide-based plant selection. The OsCc1 promoter was used to drive constitutive plasmid gene expression (Jang et al., 2002). The coding regions of AP37 and AP59 were amplified from rice (Oryza sativa) total RNA using an RT-PCR system (Promega) according to the manufacturer's instructions. Primer pairs were as follows: AP37 forward (5′-ATGGCGCCCAGAGCAGCTAC-3′) and AP37 reverse (5′-CTAGTTCTCTACCGGCGGCG-3′); and AP59 forward (5′-ATGCTGCTTAATCCGGCGTC-3′) and AP59 reverse (5′-TTAGCTCACCAGCTGCTGGA-3′). Plasmids were introduced into Agrobacterium tumefaciens LBA4404 by triparental mating, and embryogenic calli from mature seeds (cv Nakdong) were transformed as described previously (Jang et al., 1999).
Drought Treatments at the Vegetative Stage
Transgenic and NT rice seeds were germinated in half-strength Murashige and Skoog (MS) solid medium in a growth chamber in the dark at 28°C for 4 d, transplanted into soil, and then grown in a greenhouse (16-h-light/8-h-dark cycles) at 28°C to 30°C. Eighteen seedlings from each transgenic and NT line were grown in pots (3 × 3 × 5 cm; one plant per pot) for 4 weeks before undertaking the drought stress experiments. To induce drought stress, 4-week-old transgenic and NT seedlings were unwatered for 3 d followed by 7 d of watering. The numbers of plants that survived or continued to grow were then scored.
Chlorophyll Fluorescence under Conditions of Drought, High Salinity, and Low Temperature
Transgenic and NT rice seeds were germinated and grown in half-strength MS solid medium for 14 d in a growth chamber (16-h-light [150 μmol m−2 s−1]/8-h-dark cycles at 28°C). The green portions of approximately 10 seedlings were cut using a scissors prior to stress treatments in vitro. To induce low-temperature stress, the seedlings were incubated at 4°C in water for up to 6 h under continuous 150 μmol m−2 s−1 light. For high-salinity stress treatments, they were incubated in 400 mm NaCl for 2 h at 28°C under continuous 150 μmol m−2 s−1, and to simulate drought stress, they were air dried for 2 h at 28°C under continuous 150 μmol m−2 s−1 light. The Fv/Fm values were then measured as described previously (Oh et al., 2005b).
Rice 3′-Tiling Microarray Analysis
Expression profiling was conducted using the Rice 3′-Tiling microarray manufactured by NimbleGen (http://www.nimblegen.com/), which contains 27,448 genes deposited at the International Rice Genome Sequencing Project Rice Annotation Project 1 database (http://rapdb.lab.nig.ac.jp). Further information on this microarray, including statistical analysis, can be found at http://www.ggbio.com (GreenGene Biotech). Among the genes on the microarray, 20,507 are from representative Rice Annotation Project 1 sequences with cDNA/EST supports, and 6,941 genes have been predicted without cDNA/EST supports. Ten 60-nucleotide-long probes were designed from each gene starting at 60 bp ahead of the stop codon and with 10-bp shifts in position, so that 10 probes covered 150 bp within the 3′ region of the gene. In total, 270,000 probes were designed in this way (average size, 60 nucleotides) to have melting temperature values of between 75°C and 85°C. Random GC probes (38,000) were used to monitor the hybridization efficiency, and fiducial markers at the four corners (225) were included to assist with overlaying of the grid on the image.
To identify stress-inducible AP2 genes in rice, total RNA (100 μg) was prepared using 14-d-old rice leaves from plants subjected to drought, high-salinity, ABA, and low-temperature stress conditions. For the high-salinity and ABA treatments, the 14-d-old seedlings were transferred to a nutrient solution containing 400 mm NaCl or 100 μm ABA for 2 h in the greenhouse under continuous light of approximately 1,000 μmol m−2 s−1. For drought treatment, 14-d-old seedlings were air dried for 2 h under continuous light of approximately 1,000 μmol m−2 s−1. For low-temperature treatment, 14-d-old seedlings were exposed at 4°C in a cold chamber for 6 h under continuous light of 150 μmol m−2 s−1. For identification of genes up-regulated in OsCc1:AP37 and OsCa1:AP59 plants, total RNA (100 μg) was prepared from leaf tissues of 14-d-old transgenic and NT rice seedlings grown under normal growth conditions. The mRNA was purified using the Qiagen Oligotex column according to the manufacturer's instructions. For normalization, data were processed with cubic alpine normalization using quartiles to adjust signal variation between chips and with robust multi-chip analysis using a median polish algorithm implemented in NimbleScan (Workman et al., 2002; Irizarry et al., 2003). To assess the reproducibility of the microarray analysis, we repeated the experiments three times with independently prepared total RNAs and analyzed each data set statistically using one-way ANOVA.
RT-PCR and Quantitative PCR Analysis
Total RNA was prepared as reported previously (Oh et al., 2008). For the analysis of target by RT-PCR, a cDNA synthesis system (Invitrogen) was used according to the manufacturer's instructions. PCR products were amplified using primers designed with Primer Designer 4 software (Sci-ed Software). RT-PCR was carried out using the primer pairs listed in Supplemental Table S3 at a final concentration of 10 pm each and 2 μL (equivalent to 5 ng of total RNA) of cDNA as the template. PCR was performed at 95°C for 10 min, followed by 20 to 25 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 1 min. Amplified products were resolved on a 2% agarose gel. To validate our RT-PCR results, we repeated each experiment twice with independently prepared total RNA.
For quantitative real-time PCR experiments, the SuperScript III Platinum One-Step Quantitative RT-PCR system (Invitrogen) was used. For PCR, a master mix of reaction components was prepared according to the manufacturer's protocol for Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Thermocycling and fluorescence detection were performed using the Mx3000p Real-Time PCR machine (Stratagene). PCR was performed at 95°C for 10 min, followed by 20 to 25 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 1 min. To validate our quantitative PCR results, we repeated each experiment three times.
Drought Treatments in the Field for Reproductive-Stage Rice Plants
To evaluate yield components of transgenic plants under normal field conditions, three independent T5 homozygous lines of the OsCc1:AP37 and OsCc1:AP59 plants, together with NT controls, were transplanted to a paddy field at the Rural Development Administration, Suwon, Korea. A completely randomized design was employed with two replicates, each consisting of four plots of 5 m2. At 25 d after sowing, the seedlings were randomly transplanted within a 15- × 30-cm distance. Fertilizer was applied at 70:40:70 (nitrogen:phosphorus:potassium) kg ha−1 after the last paddling and 45 d after transplantation. Yield parameters were scored for 10 plants per plot and 20 plants per line.
To evaluate yield components of transgenic plants under drought field conditions, three independent T5 homozygous lines of the OsCc1:AP37 and OsCc1:AP59 plants and NT controls were transplanted to a removable rain-off shelter with a 1-m-deep container filled with natural paddy soil located at Myongji University, Yongin, Korea. Completely randomized design, transplanting distance, and use of fertilizer were employed as described above for normal field conditions. Drought stress was applied at the panicle heading stage (from 10 d before heading to 20 d after heading) by flowing water through a drain at the bottom of the container. To prevent plants from dying, we irrigated twice when the plant leaves rolled during drought stress. After exposure to drought stress conditions, the polyvinyl roofs were removed and plants were irrigated until harvesting.
When the plants grown under normal and drought conditions had reached maturity and the grains had ripened, they were harvested and threshed by hand (separation of seeds from the vegetative parts). The unfilled and filled grains were taken apart, independently counted using a Countmate MC1000H (Prince), and weighed. The following agronomic traits were scored: flowering date, panicle length, number of tillers, number of panicles, spikelets per panicle, filling rate (%), total grain weight (g), and 1,000 grain weight (g). The results from three independent lines were separately analyzed by one-way ANOVA and compared with those of the NT controls. The ANOVA was used to reject the null hypothesis of equal means of transgenic lines and NT controls (P < 0.05). SPSS version 16.0 was used to perform statistical analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK061380 (AP37) and AK073812 (AP59).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenic relationship of the rice AP2 family.
Supplemental Figure S2. Up-regulation of five target genes in OsCc1:AP37 and OsCc1:AP59 plants in comparison with NT controls.
Supplemental Table S1. Up-regulated genes in OsCc1:AP37 and OsCc1:AP59 plants in comparison with NT controls.
Supplemental Table S2. Agronomic traits of the OsCc1:AP37 and OsCc1:AP59 transgenic rice plants under normal growth and drought stress conditions in the field.
Supplemental Table S3. Primers used for RT-PCR.
Supplementary Material
Acknowledgments
We are grateful to Dr. Soon Jong Kweon at the National Academy of Agricultural Sciences, Rural Development Administration, of Korea for making critical comments on the field experiments and analyses.
This work was supported by the Ministry of Education, Science, and Technology of Korea through the Crop Functional Genomics Center (grant no. CG2111 to J.-K.K.), by the Biogreen21 Program (grant to J.-K.K.), and by the Korea Science and Engineering Foundation through the Plant Metabolism Research Center at Kyung-Hee University (grant to J.-K.K.)
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ju-Kon Kim (jukon306@gmail.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Asch F, Dingkuhn M, Sow A, Audebert A (2005) Drought-induced changes in rooting patterns and assimilate partitioning between root and shoot in upland rice. Field Crops Res 93 223–236 [Google Scholar]
- Boonjung H, Fukai S (1996) Effects of soil water deficit at different growth stage on rice growth and yield under upland conditions. 2. Phenology, biomass production and yield. Field Crops Res 48 47–55 [Google Scholar]
- Choi DW, Rodriguez EM, Close TJ (2002) Barley Cbf3 gene identification, expression pattern, and map location. Plant Physiol 129 1781–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev 12 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) DREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33 751–763 [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcription activators as an early step in cold-inducible COR gene expression. Plant J 16 433–442 [DOI] [PubMed] [Google Scholar]
- Guo ZJ, Chen XJ, Wu XL, Ling JQ, Xu P (2004) Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol Biol 55 607–618 [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7 465–471 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT (2002) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67 169–181 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47 141–153 [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Kaake V, Xhan JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127 910–917 [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106 [DOI] [PubMed] [Google Scholar]
- Jang IC, Choi WB, Lee KH, Song SI, Nahm BH, Kim JK (2002) High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol 29 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Nahm BH, Kim JK (1999) Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breed 5 453–461 [Google Scholar]
- Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, et al (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress-tolerance without stunting growth. Plant Physiol 131 516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M (2006) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 22 575–588 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45 346–350 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulated through the drought-responsive element in an ABA-dependent pathway. Plant J 30 679–689 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51 617–630 [DOI] [PubMed] [Google Scholar]
- Oh SJ, Jeong JS, Kim EH, Yi NR, Yi SI, Jang IC, Kim YS, Suh SC, Nahm B, Kim JK (2005. a) Matrix attachment region from the chicken lysozyme locus reduces variability in transgene expression and confers copy number-dependence in transgenic rice plants. Plant Cell Rep 4 145–154 [DOI] [PubMed] [Google Scholar]
- Oh SJ, Kim SJ, Kim YS, Park SH, Ha SH, Kim JK (2008) Arabidopsis cyclin D2 expressed in rice forms a functional cyclin-dependent kinase complex that enhances seedling growth. Plant Biotechnol Rep 2 227–231 [Google Scholar]
- Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK (2007) Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol J 5 646–656 [DOI] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005. b) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, Yan DQ, Du BX, Zhang JS, Liu Q, Chen SY (2003) Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis. Theor Appl Genet 107 155–161 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6 410–417 [DOI] [PubMed] [Google Scholar]
- Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59 533–551 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50 571–599 [DOI] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menéndez C, Ochogavía ME, Coll Y, Hernández I, Borrás-Hidalgo O, Thomma BPHJ, Vera P, Hernández L (2008) SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol 49 512–525 [DOI] [PubMed] [Google Scholar]
- Weigel D (1995) The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7 388–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis MCS, Kropff MJ, Maligaya AR, Tuong TP (1996) Drought-stress responses of two lowland rice cultivars to soil water status. Field Crops Res 46 21–39 [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method to reduce variability in DNA microarray experiments. Genome Biol 3 research0048.1–research0048.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Z, Zhang H, Wang XC, Huang R (2008) Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol 148 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115 35–46 [DOI] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP (2002) An AP2 domain transcription factor HvCBF1 activates expression of cold-responsive genes in barley through interaction with a (G/a)(C/t)CGAC motif. Biochim Biophys Acta 1577 63–72 [DOI] [PubMed] [Google Scholar]
- Xue GP (2003) The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33 373–383 [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D (2004) The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol 136 2862–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.