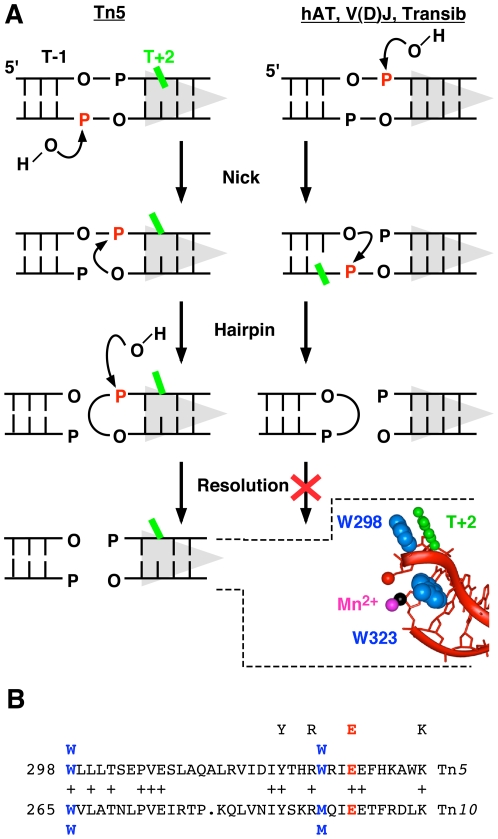
Figure 1. The hairpin and base flipping stages of transposition in different families of transposons.
A Different families of DDE transposons have hairpin intermediates of opposite polarity. Scissile phosphates are shown in red; transposon end and RSS, grey triangles. Left panel: Binding of Tn5 transposase creates a distortion in the DNA that destabilizes stacking of the T+2 base (green). The first step of the reaction is a nick to expose the 3′-OH at the end of the transposon. This facilitates flipping of the T+2 base from the helix in preparation for cleavage of the second strand by a direct transesterification reaction, generating a hairpin intermediate on the transposon end [35], [36]. Subsequently, the hairpin is resolved to yield a blunt transposon end. The insert shows the co-crystal structure of the Tn5 transposon end, with the flipped base at position +2 [1]. All of the residues of the bound transposase have been omitted except for two tryptophan residues. One acts as a probe inserted into the DNA helix, while the other provides stacking interactions to stabilize the flipped base. Right panel: In the hAT transposons and V(D)J recombination the polarity of the reaction is reversed. The first nick occurs on the top strand generating a 3′-OH on the flanking DNA end [2], [37], [38]. Transesterification yields a hairpin on the flanking DNA that is processed by the host. Residue -1 on the bottom strand is distorted and becomes permanganate sensitive after the first nick (green) [17]. B The amino acid sequence of the Tn5 transposase in the vicinity of the probe and stacking residues is aligned with the equivalent region from Tn10 transposase. The Tn5 transposase stacking tryptophan is at position 298 and a tryptophan occupies the equivalent position in Tn10 transposase. However, the Tn5 transposase probe tryptophan aligns with a methionine residue in the Tn10 transposase. The E residue of the YREK motif is also a member of the DDE triad of residues that coordinate the catalytic metal ions and are essential for catalysis.