Abstract
Prader-Willi syndrome (PWS) is caused by deficiency for one or more paternally expressed imprinted transcripts within chromosome 15q11-q13, including SNURF-SNRPN and multiple small nucleolar RNAs (snoRNAs). Balanced chromosomal translocations that preserve expression of SNURF-SNRPN and centromeric genes but separate the snoRNA HBII-85 cluster from its promoter cause PWS. A microdeletion of the HBII-85 snoRNAs in a child with PWS provides, in combination with previous data, effectively conclusive evidence that deficiency of HBII-85 snoRNAs causes the key characteristics of the PWS phenotype, although some atypical features suggest that other genes in the region may make more subtle phenotypic contributions.
Large interstitial deletions of paternal origin on chromosome 15q11–q13 are the cause of Prader-Willi syndrome (MIM176270) in ∼70% of cases. Most remaining affected individuals have maternal uniparental disomy 15, and some have imprinting defects. A number of paternally expressed genes mapping within this critical region have been suggested to have a role in the pathogenesis of PWS, including SNURF-SNRPN, which encodes the SNURF and SNRPN polypeptides in a single transcript. Identification of individuals with PWS with balanced translocations leaving the SNURF-SNRPN promoter and coding regions intact later excluded SNURF-SNRPN as a candidate gene1-4. Located within the introns of very long transcripts extending downstream of SNRPN, there are clusters of paternally expressed C/D box–containing snoRNAs that are highly expressed in the brain5,6. SnoRNAs represent an important subset of noncoding molecules with diverse functions, the best documented being site-specific covalent modifications of ribosomal RNAs and small nuclear RNAs by 2′-O-methylation5,6. The translocation cases that exclude SNURF and SNRPN as candidate genes suggest that snoRNA deficiency might cause PWS, but expression of the translocated snoRNAs in the brain could not be tested3,4.
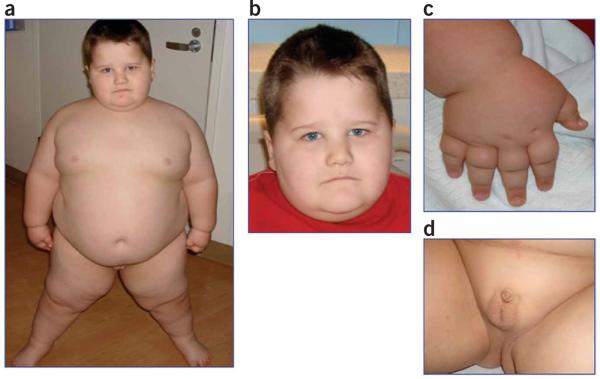
Here, we describe the characterization of a de novo microdeletion in an individual meeting the criteria for a diagnosis of PWS, showing all of seven major revised clinical criteria including neonatal hypotonia, feeding difficulties and failure to thrive during infancy, excessive weight gain after 18 months, hyperphagia, hypogonadism, global developmental delay and equivocal facial features (Fig. 1, Supplementary Table 1 and Supplementary Note online)7. He has an atypical deletion within 15q11–q13. Additional minor features include behavioral problems, sleep apnea, skin picking, speech delay, and small hands and feet relative to height (Fig. 1c, Supplementary Note and Supplementary Tables 1 and 2 online). His cognitive skills fall within the range of mild mental retardation, and he meets the Autism Diagnostic Observation Schedule (ADOS) and Autism Diagnostic Interview-Revised (ADI-R) defined criteria for autism. Atypical findings included a height at the 95th percentile and a head circumference above the normal range. The facies and hand configuration were equivocal in terms of being typical for PWS (Fig. 1). A comparison between the clinical and physical features of this individual and those of other individuals with PWS who carry balanced translocations involving 15q11-q13 is shown in Supplementary Table 1, and other possible diagnoses were considered and excluded as described in the Supplementary Note. Informed parental consent, as approved by the Baylor College of Medicine Institutional Review Board, was obtained prior to research studies.
Figure 1. Clinical features of the affected individual.
(a,b) Individual showing morbid obesity with facial features as shown. (c) Upper extremities are notable for small hands relative to body size. (d) External genitalia after laparoscopic orchiopexy at 13 months. Parental informed consent, as approved by the Baylor College of Medicine Institutional Review Board, was obtained to publish the photographs.
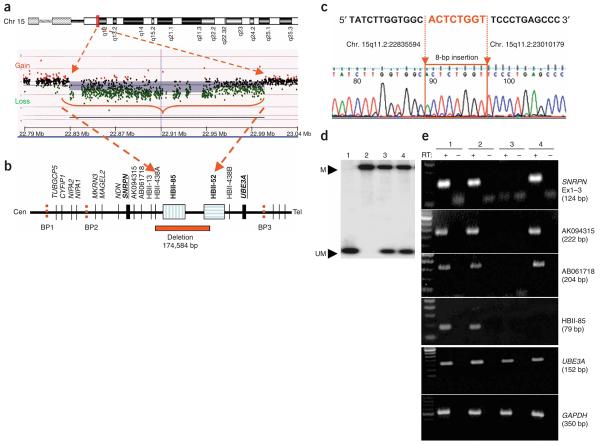
This individual was found to have a deletion in the snoRNA region at 15q11.2 (Fig. 2). Array-based comparative genomic hybridization (array CGH) using a BAC array showed a loss of copy number for two clones encompassing ∼400 kb within the 15q11–q13 PWS and Angelman Syndrome critical interval (Supplementary Fig. 1a and Supplementary Methods online). FISH studies with the specific clones suggested a weak but not absent signal (Supplementary Fig. 1b). To rule out deletion or imprinting abnormalities causing PWS, we carried out DNA methylation analysis of the PWS-imprinting center and found a normal methylation pattern (Fig. 2d). Chromosome analysis showed a normal male karyotype. A combination of high-resolution oligonucleotide-based array CGH and quantitative real-time PCR helped to define the deletion more precisely (Fig. 2a,b and Supplementary Fig. 1c,d), and predicted the deletion boundaries to be between positions 22.83 Mb (centromeric) and 23.01 Mb (telomeric). The clinical array used to make the initial diagnosis in the affected individual included 1,475 BAC clones and did not detect any other abnormalities. We identified a junction fragment of ∼2.6 kb using long-range PCR and breakpoints at position 22,835,594 (proximal) and 23,010,179 (distal) with an insertion of 8 bp using sequencing (Fig. 2b,c and Supplementary Fig. 1e); thus, the deleted segment was exactly 174,584 bp. The proximal breakpoint occurred between EST AB061718 and snoRNA HBII-438A, and the distal breakpoint was between snoRNA box 23 and 25 of the HBII-52 cluster (Fig. 2b). The deletion encompasses HBII-438A, all 29 snoRNAs comprising of the HBII-85 cluster, and the proximal 23 of the 42 snoRNAs comprising the HBII-52 cluster. The parental origin of the deletion was confirmed by a polymorphic dinucleotide repeat within the deleted interval; this showed that only a maternal allele was present in the proband, thus placing the deletion on the paternal chromosome (Supplementary Fig. 2a online). A search for genomic elements in the vicinity of the breakpoints did not show recognizable features that might contribute to a recombination-based deletion event, except for the presence of four Alu (short interspersed nuclear element) and two long interspersed nuclear elements (L1D2 and L1MB7) within 5 kb centromeric of the breakpoint.
Figure 2. Analysis of microdeletion causing PWS and expression studies.
(a) A high-resolution oligonucleotide array-CGH plot is shown with loss of a segment in 15q11.2 from position ∼22,835,000 bp to ∼23,010,100 bp (red arrows). (b) A schematic physical map of the 15q11–q13 genomic interval is shown, highlighting the deleted segment with respect to SNRPN, UBE3A and snoRNAs within the interval. (c) Sequencing of a ∼2.6-kb PCR fragment across the breakpoint revealing a deletion of 174,584 bp with an 8 bp insertion at the breakpoint. (d) Methylation analysis by DNA blot hybridization to rule out large deletion, uniparental disomy or imprinting abnormalities. Probe corresponding to SNRPN exon 1 was used for hybridization, and a normal methylation pattern was seen. Lane 1, large deletion AS; 2, large deletion PWS; 3, normal control; 4, affected individual. M, methylated maternal allele; UM, unmethylated paternal allele. (e) Expression analysis was carried out using RT-PCR of lymphoblast RNA for SNRPN, snoRNA HBII-85, ESTs AK094315 and AB061718, and UBE3A (size of PCR products is in parentheses). GAPDH was used as internal RT-PCR control. RT+, with reverse transcriptase; RT−, without reverse transcriptase. Affected individual shows lack of HBII-85 transcript in RT+ lane with presence of transcripts for all other loci tested. Lane 1, normal control; 2, large deletion AS; 3, large deletion PWS; 4, affected individual.
To determine the expression of upstream and downstream transcripts, we carried out RT-PCR using RNA from lymphoblasts of the affected individual and appropriate normal and disease controls. RT-PCR showed normal expression of SNRPN, HBII-13 (data not shown), ESTs AK094315 and AB061718, and HBII-438A/B in the proband (Fig. 2e and Supplementary Fig. 2b). UBE3A is maternally expressed in brain but is transcribed from both alleles in peripheral tissues, and expression was detected in lymphoblasts as expected. HBII-52 snoRNA expression has been shown to be absent in peripheral tissues, and no expression was seen for lymphoblast RNA from controls or the affected individual (data not shown)5,6. Expression analysis of HBII-438A gave positive results as a result of the presence of an identical copy of this snoRNA box (HBII-438B) located just distal to the HBII-52 cluster. Expression of NDN was negative as expected, as it is not expressed in lymphoblasts (Supplementary Fig. 2c).
Our data thus reveal a unique microdeletion encompassing the entire HB-II-85 cluster, HBII-438A, and a portion of the HB-II-52 cluster of snoRNAs. Six PWS cases with balanced translocation breakpoints within or downstream of the SNRPN coding exons suggest an important role for the paternally expressed snoRNAs located between UBE3A and SNRPN1-4,8-11. The HBII-85 snoRNAs are more likely candidates for causing PWS than the HBII-52 cluster, because two families with a microdeletion of the entire HBII-52 cluster did not manifest an obvious phenotype with paternal inheritance of the deletion8,12. Most of the individuals with the translocation have typical PWS phenotypes, although there are exceptions3. Expression of the snoRNAs in the brain cannot be predicted in translocation cases, because the snoRNA genes are separated from their normal promoter. One individual with the translocation lacked infantile hypotonia, neonatal feeding difficulties and typical facies2. This could be explained by some level of transcription of the transclocated snoRNA loci in the brain of this individual. Although the paternally expressed NDN (necdin) gene has been associated with PWS, this finding is based primarily on mouse data, and data supporting a role for NDN in human PWS are lacking. Hence, the data discussed above as ruling out involvement of SNRPN equally argue against a major role for the NDN gene in causing the PWS phenotype. The HBII-52 snoRNA has been shown to regulate alternative splicing of a serotonin receptor (5-HT2CR)13. Although it has been suggested that the effect on splicing explains why individuals with PWS respond to selective serotonin reuptake inhibitor treatment and points to defects in the serotoninergic system as a contributing cause of PWS13, this interpretation is speculative, and there is no conclusive evidence that paternal deficiency for HBII-52 contributes to the PWS phenotype8,12,13. Although we cannot exclude the possibility that partial deficiency of HBII-52 modifies the phenotype produced by paternal deficiency of HBII-85, the data are now notably in favor of the interpretation that paternal deficiency of HBII-85 causes the key manifestations of the PWS phenotype, although some atypical features suggest that other genes in the region may make lesser phenotypic contributions (for example, the tall stature of the individual in this study, and hypopigmentation seen with large deletions).
Recently, two mouse models were generated with targeted deletions of the MBII-85 snoRNA cluster14,15. Although neonatal lethality was variable up to 15% and background specific in one model15, both mouse models showed decreased activity and/or hypotonia at birth and postnatal growth retardation, as is typical for other PWS mouse models with larger deletions. As in the case of other PWS mouse models, these mice were not obese or infertile, implying species-specific differences. One mouse model was described as having deficiency in motor learning, increased anxiety and hyperphagia without obesity14. These mice recapitulated an important subset of the PWS phenotype, providing additional evidence for the hypothesis that absence of the snoRNAs contributes to disease pathogenesis.
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grants HD-037283 and M01-RR00188 (General Clinical Research Center), HD-024064 (Mental Retardation and Developmental Disabilities Research Center) and RR-019478 (Rare Disease Clinical Research Consortia). We thank J. Bressler for critical reading of the manuscript.
Footnotes
Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Schulze A, et al. Nat. Genet. 1996;12:452–454. doi: 10.1038/ng0496-452. [DOI] [PubMed] [Google Scholar]
- 2.Wirth J, et al. Hum. Mol. Genet. 2001;10:201–210. doi: 10.1093/hmg/10.3.201. [DOI] [PubMed] [Google Scholar]
- 3.Schüle B, et al. BMC Med. Genet. 2005;6:18. doi: 10.1186/1471-2350-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallagher RC, et al. Am. J. Hum. Genet. 2002;71:669–678. doi: 10.1086/342408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Runte M, et al. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 6.de los Santos T, et al. Am. J. Hum. Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunay-Aygun M, et al. Pediatrics. 2001;108:e92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- 8.Runte M, et al. Hum. Genet. 2005;116:228–230. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, et al. Hum. Mol. Genet. 1996;5:517–524. doi: 10.1093/hmg/5.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuslich CD, et al. Am. J. Hum. Genet. 1999;64:70–76. doi: 10.1086/302177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conroy JM, et al. Am. J. Hum. Genet. 1997;61:388–394. doi: 10.1086/514852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger J, et al. Am. J. Med. Genet. 2002;111:233–237. doi: 10.1002/ajmg.10498. [DOI] [PubMed] [Google Scholar]
- 13.Kishore S, et al. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 14.Ding F, et al. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skryabin BV, et al. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]