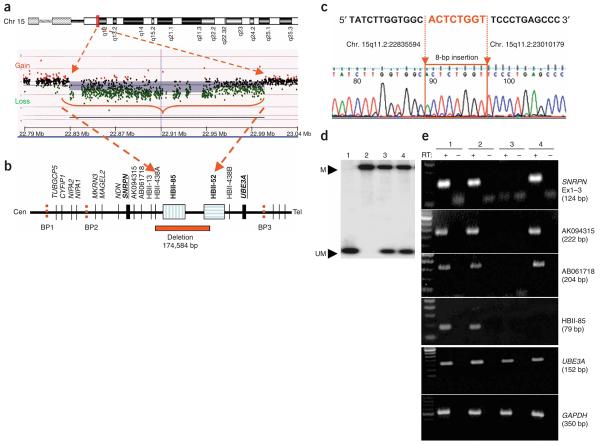
Figure 2. Analysis of microdeletion causing PWS and expression studies.
(a) A high-resolution oligonucleotide array-CGH plot is shown with loss of a segment in 15q11.2 from position ∼22,835,000 bp to ∼23,010,100 bp (red arrows). (b) A schematic physical map of the 15q11–q13 genomic interval is shown, highlighting the deleted segment with respect to SNRPN, UBE3A and snoRNAs within the interval. (c) Sequencing of a ∼2.6-kb PCR fragment across the breakpoint revealing a deletion of 174,584 bp with an 8 bp insertion at the breakpoint. (d) Methylation analysis by DNA blot hybridization to rule out large deletion, uniparental disomy or imprinting abnormalities. Probe corresponding to SNRPN exon 1 was used for hybridization, and a normal methylation pattern was seen. Lane 1, large deletion AS; 2, large deletion PWS; 3, normal control; 4, affected individual. M, methylated maternal allele; UM, unmethylated paternal allele. (e) Expression analysis was carried out using RT-PCR of lymphoblast RNA for SNRPN, snoRNA HBII-85, ESTs AK094315 and AB061718, and UBE3A (size of PCR products is in parentheses). GAPDH was used as internal RT-PCR control. RT+, with reverse transcriptase; RT−, without reverse transcriptase. Affected individual shows lack of HBII-85 transcript in RT+ lane with presence of transcripts for all other loci tested. Lane 1, normal control; 2, large deletion AS; 3, large deletion PWS; 4, affected individual.