Fig. 6. CLASPs preferentially bind to MTs originated at the Golgi.
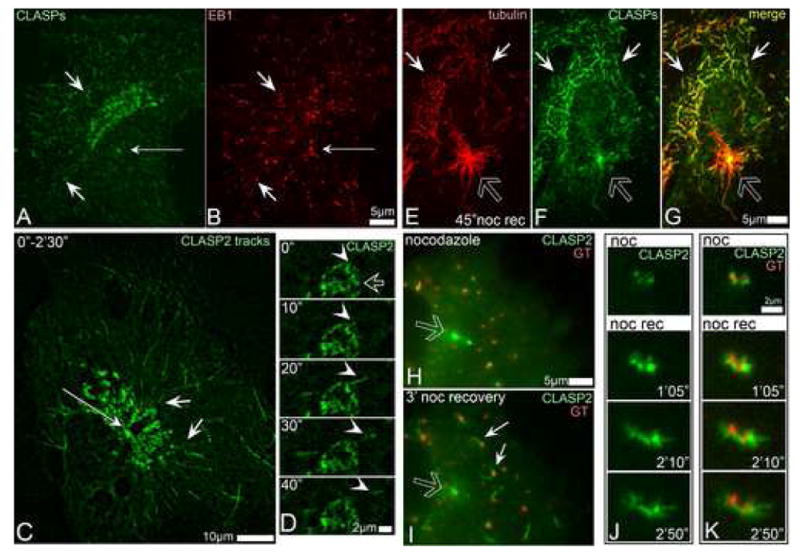
A, CLASPs (green) in RPE1 cells are associated with MT tips close to the Golgi (short arrows), but not around the centrosome (thin arrow). B, EB1 (red) at the MT tips in the cell shown in G. Immunostaining. C, Overlaid live recording of GFP-CLASP2-expressing RPE1 cell within 2.5 min. CLASP2-associated MT tracks (short arrows) radiate from the TGN but not form the centrosome (thin arrow). D, Live sequence illustrating formation of GFP-CLASP2-decorated MT (arrowhead) at CLASP-rich TGN (hollow arrow) in untreated cell. CLASP2 coats whole newly formed MT (10″-20″) but remains only at the tip at a later stage (30″-40″). E-G, A cell fixed 45″ after nocodazole washout. E, tubulin (red). F, CLASPs (green). G, Merge. CLASPs localize to non-centrosomal (white arrows) but not to the centrosomal (hollow arrow) MTs. H-I, GFP-CLASP2 (green) localizations before and after nocodazole washout. In nocodazole (left) CLASP2 associate with Golgi stacks (mCherry-GT, red) and with the centrosome (hollow arrow). 3′ after nocodazole removal (right) CLASP2 is detected at the Golgi-associated MTs (white arrows) but not around the centrosome (hollow arrow). J-K, Video frames illustrating formation of GFP-CLASP2-coated MTs at GFP-CLASP2-enriched Golgi stacks in nocodazole washout. J, GFP-CLASP2. K, GFP-CLASP2 (green) and mCherry-GT (red). Time after nocodazole removal is shown.
