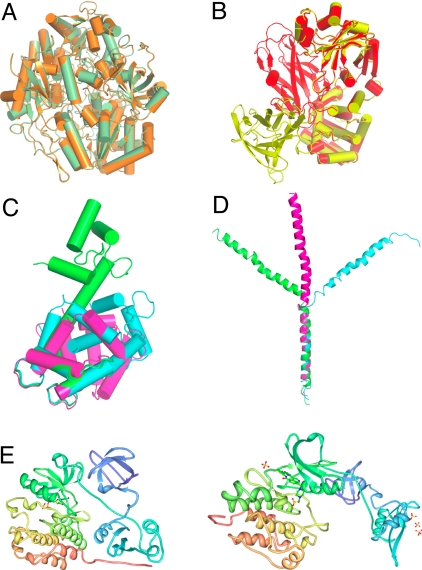
Fig. 7.
Examples of proteins representing different ‘mobility classes’ found in the PDB. (A) cluster 48 representing a large enzyme transhydroxylase (1,236 amino acids) with a single conformational state. (B) Cluster 633 representing diphtheria toxin with a single hinge that represents a movement of the entire domain shown in yellow and red models. (C) Cluster 8,791 representing calmodulin that has the entire family of different conformational states represented. Three of these states are depicted; green an open state, blue half closed state, and purple the completely closed state. (D) Cluster 11,575 representing a fragment of apolipoprotein A. Three conformational states are represented out of many available in 2 independent crystal structures. (E) The Src kinase representing the largest conformational change detected in the PDB models represented in our database that comprises 23.7 Å RMSD between models 2src and 1y57.