Table 1. Crystallographic statistics.
Values in parentheses are for the outermost shell.
| RNase A–U5P | RNase A–UDP | |
|---|---|---|
| Space group | C2 | C2 |
| Unit-cell parameters (Å, °) | a = 100.035, b = 32.299, c = 72.475, α = 90.00, β = 90.91, γ = 90.00 | a = 100.003, b = 32.337, c = 72.299, α = 90.00, β = 90.72, γ = 90.00 |
| Matthews coefficient (Å3 Da−1) | 2.10 | 2.09 |
| Resolution (Å) | 30.0–1.40 (1.42–1.40) | 30.0–1.40 (1.42–1.40) |
| Reflections measured | 414856 | 297437 |
| Unique reflections | 44340 (2291) | 45026 (2290) |
| Rmerge† | 0.106 (0.263) | 0.044 (0.111) |
| Completeness (%) | 95.3 (99.8) | 97.8 (100.0) |
| 〈I/σ(I)〉 | 34.5 (4.1) | 29.0 (8.2) |
| Rcryst‡ | 0.208 (0.217) | 0.188 (0.186) |
| Rfree§ | 0.254 (0.273) | 0.225 (0.270) |
| No. of solvent molecules | 358 | 367 |
| R.m.s. deviation from ideality | ||
| In bond lengths (Å) | 0.009 | 0.009 |
| In angles (°) | 1.4 | 1.4 |
| Average B factor (Å2) | ||
| Protein atoms (mol A/mol B) | 19.9/19.9 | 17.6/16.3 |
| Solvent molecules | 32.4 | 31.7 |
| Ligand atoms (mol A/mol B/mol C/mol D) | 24.2/20.9/24.7 | 26.5/22.7/24.1/28.2 |
R
merge = 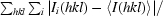
 , where I
i(hkl) and 〈I(hkl〉 are the ith and the mean measurements of the intensity of reflection hkl.
, where I
i(hkl) and 〈I(hkl〉 are the ith and the mean measurements of the intensity of reflection hkl.
R
cryst = 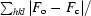
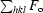 , where F
o and F
c are the observed and calculated structure-factor amplitudes of reflection hkl, respectively.
, where F
o and F
c are the observed and calculated structure-factor amplitudes of reflection hkl, respectively.
R free is the same as R cryst but for a randomly selected 5% subset of reflections not used in the refinement (Brünger, 1992 ▶).
