Abstract
Glioblastoma multiforme (GBM) remains refractory to conventional therapy. CD133+ GBM cells have been recently isolated and characterized as chemo-/radio-resistant tumor-initiating cells and are hypothesized to be responsible for post-treatment recurrence. In order to explore the molecular properties of tumorigenic CD133+ GBM cells that resist treatment, we isolated CD133+ GBM cells from tumors that are recurrent and have previously received chemo-/radio-therapy. We found that the purified CD133+ GBM cells sorted from the CD133+ GBM spheres express SOX2 and CD44 and are capable of clonal self-renewal and dividing to produce fast-growing CD133− progeny, which form the major cell population within GBM spheres. Intracranial injection of purified CD133+, not CD133− GBM daughter cells, can lead to the development of YKL-40+ infiltrating tumors that display hypervascularity and pseudopalisading necrosis-like features in mouse brain. The molecular profile of purified CD133+ GBM cells revealed characteristics of neuroectoderm-like cells, expressing both radial glial and neural crest cell developmental genes, and portraying a slow-growing, non-differentiated, polarized/migratory, astrogliogenic, and chondrogenic phenotype. These data suggest that at least a subset of treated and recurrent GBM tumors may be seeded by CD133+ GBM cells with neural and mesenchymal properties. The data also imply that CD133+ GBM cells may be clinically indolent/quiescent prior to undergoing proliferative cell division (PCD) to produce CD133− GBM effector progeny. Identifying intrinsic and extrinsic cues, which promote CD133+ GBM cell self-renewal and PCD to support ongoing tumor regeneration may highlight novel therapeutic strategies to greatly diminish the recurrence rate of GBM.
Electronic supplementary material
The online version of this article (doi:10.1007/s11060-009-9919-z) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma, Cancer stem cells, Self-renewal, Radial glial cells, Neural crest cells, Expression microarray
Introduction
Glioblastoma multiforme (GBM, World Health Organization/WHO grade IV) remains virtually incurable despite extensive surgical excision and post-operative adjuvant radio/chemotherapy. Currently, most anti-cancer therapies aim to eliminate rapidly proliferating tumor cells; thus, the novel discovery of rare and radioresistant CD133+ GBM stem cells possessing the enhanced ability to repopulate tumors by multiple laboratories [1–5] provide a potential model to explain the inability to eradicate malignant GBM tumors. Tumor recurrence after treatment may mimic the scenario of post-injury tissue repair and regeneration. Many adult tissues undergo renewal after aging or injury, and hence require a new supply of cells originating from specialized tissue stem cells with the capability to undergo self-renewal, asymmetric cell division, and multipotent differentiation to repair aged cells or damaged tissue [6–9]. Stem cells often reside in stem cell niches that provide a specialized environment to maintain and regulate their properties and activity [10, 11]. The cellular hierarchy of tissue regeneration by resident stem cells has been described in the hematopoietic system, gut, and skin [12–14]. Tissue stem cells are most often slow-cycling and give rise to daughter transient amplifying cells (TAC) that make up the majority of the proliferative cell population in the tissues, and eventually differentiate into non-proliferative cells of a particular tissue type [15, 16]. The studies of airway injury/repair in animal model indicated that airway stem cells will only be induced to self-renewal when an abundant number of TAC are depleted [17, 18], and the elimination of the progenitor and stem cell pools has a consequent failure of tissue regeneration [19]. Thus, at the functional level, CD133+ GBM stem cells behave in ways that are similar to tissue stem cells; CD133+ GBM stem cells can self-renew and reconstitute the original tumor tissue when grafted into mice [1–5]. Cancer stem cells possess a multi-lineage differentiation capacity support for the hypothesis that cancer hierarchy is a result of developmental diversity among cancer cells in different states of differentiation [20–22]. However, it is plausible that multiple genetic and/or epigenetic instability that take place within tumor stem cells might prevent progeny from undergoing non-proliferative terminal differentiation, leading to uncontrolled tumor growth [23–25].
To access genes and pathways potentially associated with malignant features of GBM tumors, we recently compared the genome-wide transcription profile of GBM tumors with that of normal brain tissue and lower-grade astrocytoma [26–28]. Besides those genes associated with inflammation, coagulation, angiogenesis, and tissue remodeling, a series of genes linked with neural stem cell (NSC), mesenchymal stem cells (MSC) and skeletal/cartilage development, was determined. It thus implicates that a tissue regeneration-like reaction is constitutively activated within GBM tumor situ. The molecular profiles of tumor samples obviously do not reflect those of the CD133+ cancer stem cell population, which only forms a small fraction of the whole tumor tissue samples. In this study, we characterized CD133+ GBM stem cells purified from the passaged CD133+ GBM sphere cultures established from recurrent GBM tumors that had previous treatment. Our results indicated that these CD133+ GBM cells have an unlimited ability to repopulate tumor spheres in cultures and are capable of reconstituting a tumor in mouse brain that displays the key histopathologic features of malignant GBM tumor. Molecular profile analysis revealed CD133+ GBM cells possess neuroectodermal-like cell properties endowed with mesenchymal differentiation and astrogliogenic potentials. Additionally, a list of overexpressed genes characterized a quiescent-like state, implying that CD133+ GBM stem cells may be clinically indolent prior to entering the proliferative phase of the cell cycle to attain their malignant phenotype [29].
Materials and methods
Culture of primary GBM cells and tumor spheres
The tumor specimens were obtained from patients who underwent surgery at University of California at Los Angeles (UCLA) Medical Center. All samples were collected under protocols approved by the UCLA Institutional Review Board. The histopathologic typing and tumor grading were done by one neuropathologist according to the WHO criteria. Tumors were enzyme-digested and washed, followed by red blood cell lysis of the pellet. Cells were cultured in a serum-free NSC medium containing DMEM/Ham’s F-12 (Mediatech, Manassas, VA) supplemented with 20 ng/ml human recombinant epidermal growth factor (EGF, Sigma-Aldrich, St. Louis, MO), 20 ng/ml basic fibroblast growth factor (FGF, Chemicon, Billerica, MA), 10 ng/ml leukemia inhibitory factor (LIF, Chemicon), and 1× B27 without vitamin A (Invitrogen, Carlsbad, CA). In some cases, cells were cultured in DMEM/Ham’s F-12 supplemented with 5% fetal bovine serum for 1–2 passage followed by switching into NSC culture condition as previously reported [4]. The D431 spheres were derived from a patient with primary/de novo GBM and S496 spheres were derived from a patient with secondary/progressive GBM [27]. Both tumors received radiation and chemotherapy prior to their recurrence and re-operation. GBM sphere cultures were split with acutase weekly (Sigma-Aldrich, St. Louis, MO) and replaced with fresh media every other day.
Real-time quantitative (qt) and semi-qt reverse transcriptase polymerase chain reaction (RT-PCR) analysis
Real-time qtRT-PCR and semi-qtRT-PCR analysis were performed to verify the expression of selected genes in CD133+ GBM cells and patient tumors. Samples were subjected to total RNA extraction with RNeasy kit (QIAGEN, Valencia, CA) and reverse transcription by using a Taqman RT Reagent Kit (Applied Biosystems, Foster City, CA). Two microgram of purified total RNA was used as template in RT and cDNA synthesis was done for 1 cycle at 50°C for 30 min and 94°C for 2 min. Real-time qtRT-PCR was carried out with MJ Opticon PCR Analyzer (MJ Research, Inc., Waltham, MA) using SYBR Green PCR Core Reagents (Applied Biosystems). The reactions were cycled 30 times [50°C for 2 min and 95°C for 10 min (94°C for 15 s, 58–60°C for 1 min, and 72°C for 1 min) × 30 cycles] and the fluorescence was measured at the end of each cycle to construct amplification curves. A melting curve was done to verify the specificity of PCR products. Quantitation of transcripts was calculated based on a titrated standard curve co-run in the same experiment and calibrated with the expression level of housekeeping gene (β-actin). The semi-quantitative RT-PCR analysis was performed, using 5 μl cDNA equivalents to 100 ng total RNA. The PCR reaction cycles were carried out as described above. After amplification, PCR products (5 μl) were electrophoresed on 2% agarose gel and visualized under ultraviolet light after SYBR Green staining. Primer 3 Input (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) was used to selected primers and nonredundant specific primer sequences was verified using National Center for Biotechnology Information BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The primer sequences and expected size of amplified PCR products are listed at supplementary Table 8.
Cell proliferation assay
The proliferative activity of pre-sorted and post-sorted CD133+ and CD133− GBM cells was determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS/PMS) colorimetric assay (Promega, Madison, WI) according to the manufacturer’s instructions. Cells were seeded into 96-well tissue culture plates at a density of 10,000 cells per well in triplicate in NSC selecting media and incubated for 24 h. The optical density was measured at 490 nm after 4-h incubation with MTS/PMS reagent.
Loss of heterozygosity analysis
Genomic DNA from sorted CD133+ GBM cells was amplified, labeled, and hybridized under the manufacturer’s recommended conditions using the GeneChip Human Mapping 10 K Array XbaI 131 (Affymetrix, Santa Clara, CA). Raw allele scores were processed using Affymetrix GeneChip Chromosome Copy Number Tool 1.1 to estimate genome-wide chromosomal gains and losses.
Animal studies and preparation of paraffin slides and frozen sections
Tumorigenicity of GBM cells was determined by injecting the cells orthotopically. Six-week-old female or male Beige/SCID mice were anesthetized and positioned into a stereotactic frame. A burr hole was made using a Dremel drill approximately 3 mm lateral and 1 mm posterior to the intersection of the coronal and sagittal sutures (bregma). Cells were injected using a Hamilton syringe at a depth of 3 mm in a volume of 2 μl. Animals were sacrificed when any sign of neurological symptoms and morbidity/moribundity was observed. Brain tissue were immediately removed and fixed in 10% formalin for 24 h and then transferred to 70% ethanol. Mouse brains were embedded in paraffin in an automatic tissue processor. Brains were sectioned at 5-μm thickness and were mounted on microscope slides. Brain tissue for frozen sections was placed in O.C.T. embedding medium (Tissue-Tekm, Miles Inc.). The sample tray was briefly dipped in liquid nitrogen and was sectioned at 5-μm thickness in a −20°C cryostat and air-dried. Slides were then stored at −70°C until used for hematoxylin and eosin (H–E) stain and immunohistochemical analysis.
Immunocytochemical, histopathological, and immunohistochemical analysis
The immunocytochemical analysis was performed on GBM spheres seeded on an eight-chamber culture slide in the presence of FGF/EGF/LIF for 48 h. Cells were washed, fixed in 4% paraformaldehyde and subjected to immunofluorescent staining. The following primary antibodies were used: CD133 (1:100, Abcam, Cambridge, MA), nestin (1:200, Chemicon, Temecula, CA), SOX2 (1:400; R&D System, Minneapolis, MN), CD105 (3 μg/ml, R&D System), YKL-40 (1:50, Quidel, San Diego, CA) and collagen type I (1:50, Santa Cruz Biotech, Santa Cruz, CA). After washing, cells were incubated with rhodamine red or Alexa Fluor 488-conjugated goat anti-mouse IgG or goat anti rabbit IgG (1:200, Invitrogen) and counterstained with Hoechst 33342 (Invitrogen) to identify all nuclei. Histopathological analyses were performed on frozen section or paraffin slides stained with H–E staining as per standard technique. Immunohistochemical staining was performed on frozen-section slides. Slides were subject to a 1-h blocking step followed by the application of primary antibody or control antibody for 1 h at room temperature. The following primary antibodies were used: CD31 (1:100, Biocare Medical, Concord, CA), CD133, YKL-40 (1:100), and SOX2 (1:100). The immunodetection was performed using Vectastain ABC Standard kit and Vector NovaRED (Vector Laboratories, Burlingame, CA).
Fluorescence-activated cell sorting analyses and purification of CD133+ GBM cells
Fluorescence-activated cell sorting analyses (FACS) analyses were employed to determine the percentage of cells expressing stem cell markers. Dissociated cells were stained with the following antibodies for 30 min at 4°C: anti-CD133-APC (Miltenyi Biotech, Auburn, CA), anti-CD44-FITC (Caltag Laboratories, Burlingame, CA), anti SOX2 (indirect staining, using Alexa Fluor 488-conjugated goat anti-mouse IgG) and fluorescence-conjugated isotype IgG controls. 10,000 events were collected in each analysis. The analyses were performed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) and ≥10,000 events were collected in each analysis. To purify CD133+ GBM cells, dissociated cells were immunostained with anti-CD133-APC under the sterile condition. The CD133+ and CD133− cells were sorted and collected on a BD FACSAria™ II cell sorter at 70 psi using a 70-μm nozzle. The purity of post-sorted cells was determined.
Microarray procedures and data analysis
Molecular profiling and analysis were performed as described [27]. Briefly, cDNA was generated and converted to cRNA probes using standard Affymetrix protocols and hybridized to Affymetrix GeneChip U133 Plus 2.0 Array. The chips were scanned using the GeneArray scanner (Affymetrix). The CEL files generated by the Affymetrix Microarray Suite version 5.0 were converted into DCP files using the DNA-Chip Analyzer (dChip 1.3; http://biosun1.harvard.edu/complab/dchip/). The DCP files were globally normalized, and gene expression values were generated using the dChip implementation of perfect-match minus mismatch model-based expression index. To avoid inclusion of low-level and unreliable signals, the higher signal needed to exceed 100 and be called present by MAS 5.0 in >30% of the samples. All group comparisons were performed in dChip.
Gene annotation
Functional annotation of individual gene was obtained from NCBI/Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez), the published literature in PubMed Central (NCBI/PubMed), Online Mendelian Inheritance in Man (NCBI/OMIM), Source database (http://source.stanford.edu/cgi-bin/source/sourceSearch), Protein knowledgebase (UniProtKB) (http://beta.uniprot.org/), and Information Hyperlinked over Proteins (http://www.ihop-net.org/UniPub/iHOP/). Functional categorization of expression-based clusters based on gene ontology (GO) was performed using a web tool dChip v1.3 software (http://www.hsph.harvard.edu/~cli/complab/dchip/). After the hierarchical clustering was performed on genes, dChip searches all branches with at least four functionally annotated genes to assess whether a local cluster is enriched by genes having a particular function with GO term.
Results
GBM spheres contain a minor population of CD133+ cells and express neural and mesenchymal stem cell-associated markers
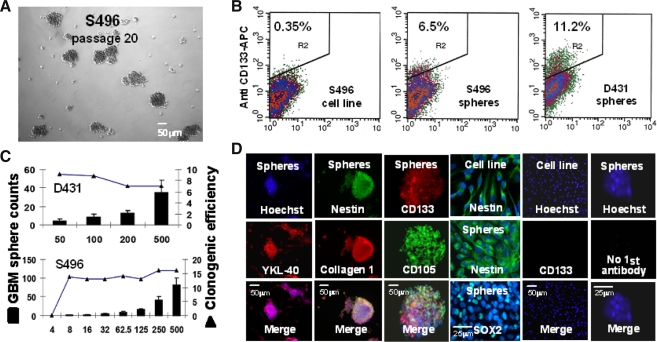
Five GBM sphere cultures were initiated under NSC-selective culture conditions; two of them (D431 and S496) were expandable, and were used for current study. GBM spheres were dissociated, clonally replated to prevent cells from forming aggregates (4 × 104 cells/7 ml/10 cm dish), and passaged weekly. Most notably, individual cells dissociated from spheres showed distinct proliferative potentials; some form abortive colonies, whereas others form larger colonies varying in size (Fig. 1A), suggesting that cells which formed spheres, are heterogeneous. When compared to autologous GBM cell cultures growing in serum-containing media, a more differentiated state, in which a negligible percent of cells express CD133 (<0.5%), 7–10% CD133+ GBM cells could be detected in GBM sphere cultures, using FACS analysis (Fig. 1B). Moreover, sphere formation analysis by limiting dilution assay revealed 7–15% clonogenic efficiency (Fig. 1C), indicating that the majority of cells within spheres are not sphere-reinitiating cells. We then test whether these bulk CD133+ GBM spheres that contain majority of CD133− GBM cells express GBM tumor-associated genes that are linked with NSC and MSC and their cell lineages as identified in GBM tumors [26–28]. Indeed, immunostaining analysis revealed several NSC- and MSC-associated markers, including SRY (sex-determining region Y)-box 2 (SOX2), nestin, YKL-40, collagen Type I, and CD105 (endoglin) were determined (Fig. 1D). RT-PCR analysis was used to confirm the expression of additional GBM markers, including maternal embryonic leucine zipper kinase (MELK), platelet-derived growth factor receptor-alpha (PDGFR-a), SOX4, and musashi homolog 1 (MSI1) (data not shown), demonstrating that cultured CD133+ GBM spheres express molecular markers of stem-like GBM tumors.
Fig. 1.
CD133+ glioblastoma (GBM) sphere culture derived from treated and recurrence GBM tumors express neural and mesenchymal stem cell-associated genes. A Under neural stem cell (NSC)-selective conditions, passaged and dissociated GBM spheres can generate single cells, small spheres, and large spheres (>50 cells), indicating tumor spheres consist of progeny with different proliferative potentials. Scale bar = 50 μm. B Propagated GBM sphere cultures contain ~7% to 10% of the CD133+ GBM cells determined by flow cytometry analysis. C The clonogenic efficiency of dissociated CD133+ GBM spheres assayed by the limiting dilutions relatively correlates to the % of the CD133+ cells determined in the GBM spheres. D CD133+ GBM spheres express neural and mesenchymal/chondrogenic-associated genes as indicated, determined by immunocytochemical analysis. Scale bar = 25 or 50 μm, as indicated
CD133+ GBM spheres were generated and maintained by CD133+ GBM cells through self-renewal and proliferative cell division
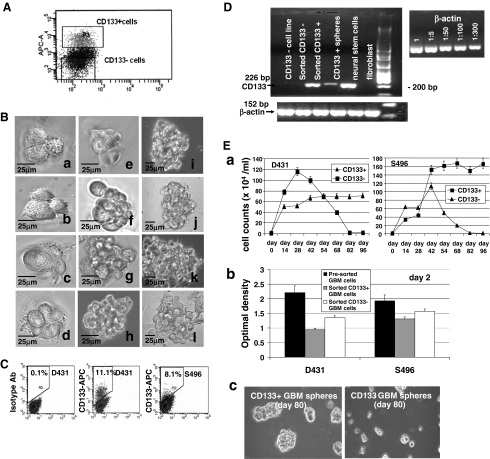
To determine whether CD133+ GBM cells are sphere-reinitiating cells, and responsible for generating CD133− GBM progeny within spheres, CD133+ GBM cells were sorted from dissociated CD133+ sphere cultures using specific CD133 antibody and FACS analysis (Fig. 2A). The purity of post sorted CD133+ cells ranged from 92% to 97%. Purified CD133+ cells were seeded in 96-well plates at the clonal density by limiting dilution. Notably, daughter cells divided from a single CD133+ GBM cell grow rapidly and gradually pile up to form GBM spheres. Moreover, populated cells are morphologically heterogeneous revealed by differences in cell size and the formed spheres showed variation in shape (Fig. 2B) [30]. These proliferative dividing cells are mostly CD133− cells as evident by the determination of ~90% CD133− GBM cells in the expanded spheres (Fig. 2C). Meanwhile, ~10% CD133+ GBM cells were determined in single cell-initiated spheres, indicating CD133+ GBM cells could clonally self-renew, but apparently be maintained in a slow-dividing status distinct from CD133− progeny. RT-PCR analysis also provided evidence for the presence of proliferative CD133− daughter cells within the growing spheres, which showed a decreased level of CD133 transcripts compared to that of purified CD133+ GBM cells (Fig. 2D). These data thus demonstrate that CD133+ GBM cells are capable of clonal self-renewal and giving rise to fast-growing CD133− daughter cells.
Fig. 2.
CD133+ GBM cells are sphere-reinitiating cells capable of undergoing clonal self-renewing and proliferative cell division in order to repopulate spheres. A Purification of CD133+ GBM cells from bulk CD133+ sphere cultures using specific anti-CD133 antibody and fluorescence-activated cell sorter. B A single CD133+ GBM cell can undergo proliferative cell division to generate daughter cells that are morphologically heterogeneous as revealed by cell size. Scale bar = 25 μm. C GBM spheres initiated by a single CD133+ GBM cell contain ~10% CD133+ GBM cells as determined by flow cytometry analysis. D RT-PCR analysis showed that purified CD133+ GBM cells overexpress CD133 transcripts compared with GBM spheres. Sorted CD133− GBM cells, serum-cultured GBM cell lines, and fibroblasts do not express CD133 transcripts. Normal neural stem cells served as positive control cells, show a strong signal for CD133. Beta-actin was served as an internal loading control. E The growth expansion assays indicated that GBM sphere cultures initiated by CD133+ GBM daughter cells, not CD133− GBM daughter cells, can be propagated for indefinite passages. (a) Cells were seeded in 6-well plates at a cell density of 104 cells per well in triplicates. Cells were counted approximately every 2 weeks and reseeded at the same cell density. (b) Short-term proliferation assay performed in day 2 cultures indicated that freshly sorted CD133+ GBM cells exhibited less proliferative activity compared to the that of CD133− GBM cells sorted from the same sphere culture, as determined by MTS/PMS colorimetric assay. Bars represent the mean ± standard error of triplicate wells. (c) CD133+ GBM cells, not CD133− GBM cells, sorted from the same CD133+ GBM sphere cultures can repopulate GBM spheres for indefinite passages
To test whether CD133+ cells are responsible for the continuous propagation of GBM sphere in cultures, we compared the growth expansion of purified CD133+ cell-initiated cultures with that of cultures initiated by the purified CD133− progeny (Fig. 2E). In order to ensure that CD133+ cells were completely removed from the CD133− fraction, post-sorted CD133− cells were subjected to a second round of cell sorting. The purified CD133+ and CD133− daughter cells sorted from the same CD133+ GBM sphere cultures initiated by the CD133+ GBM cells were respectively seeded in triplicate in 6-well plates at the cell density of 104 cells/2 ml/well. Cell counting was performed biweekly. Most notably, the growth initiation of cell cultures seeded by purified CD133+ GBM cells (40–50 × 104 cells/ml in first cell counting) was delayed compared to that of purified CD133− GBM cell-seeded culture (70–80 × 104 cells/ml), which showed an enhanced growth rate in the early passages (Fig. 2E, a). This determination was verified by a short-term (24 h) proliferation assay on day 2 after sorting, which also indicated a slower growth rate of freshly purified CD133+ GBM daughter cells compared to CD133− GBM daughter cells sorted from the same sphere cultures (Fig. 2E, b). We however, observed a lesser difference in proliferative activity between CD133+ S496− and CD133− S496 cell-seeded cultures compared to the growth differences between CD133+ and CD133− D431 cell-seed cultures. This may be due to purified CD133+ S496 GBM cell culture being able to drop from ≥95% purity to 5–10% within ~3 to 5 days, whereas it will take ~2 weeks to drop to ~10% to 15% in D431 cells (data not shown). Nevertheless, the pre-sorted GBM cells from the dissociated CD133+ GBM spheres showed a better proliferative activity than sorted CD133+ or CD133− cells in either cases (Fig. 2E, b). The growth of CD133− GBM cell-initiated cultures gradually dropped after repeated passaging in contrast to that of CD133+ cell-initiating cultures, which showed a stable expansion (Fig. 2E, a, c). These results thus indicate that CD133+ GBM cells have the capacity for unlimited self-renewal, which is required for a long-term propagation of D431 and S496 GBM spheres in cultures.
Cells spontaneously migrate out of GBM spheres and form the surrounding monolayer
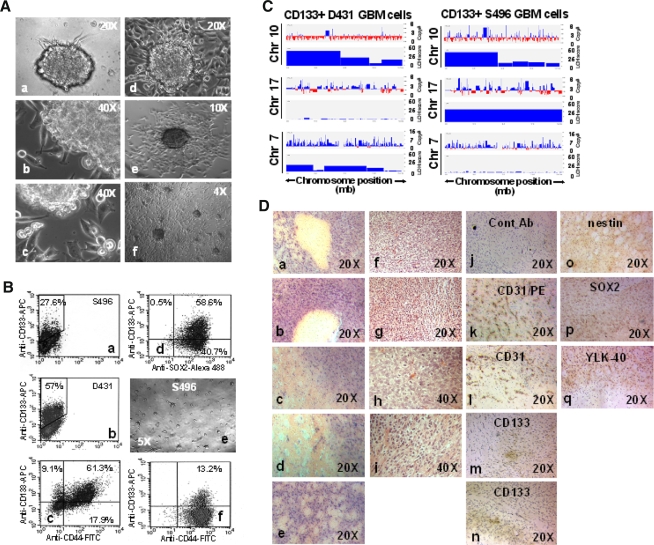
It was reported that neural precursor cells migrating out of neurospheres in cultures and outgrowing into a monolayer [31, 32]. We have also observed a similar in vitro characteristic in GBM sphere cultures. GBM cells can spontaneously migrate radially outward from the semi-adherent and flattened GBM sphere bodies, resulting in a rim of monolayer cells surrounding the spheres (Fig. 3A, a–e). Eventually, these migrating cells outgrow into an adherent monolayer that spread out over the surface of the culture dish (Fig. 3A, f). Unexpectedly, these cultures contain a higher percentage of CD133+ GBM cells (15–30% for S496 and 50–70% for D431) than non-adherent sphere cultures (Fig. 3B, a, b). The majority of CD133+ GBM cells co-express SOX2 and CD44 (Fig. 3B, c, d) as those of CD133− progeny in same cultures, indicating that CD133+ GBM cells sharing some of their surface markers with their immediate progeny grew in the same cultures. Since no additional factors were added into the culture to influence the behavior of cells, such a cell migration may be an intrinsic property that reflects inherently migratory properties of the GBM tumor of origin, which may confer a infiltrative nature of GBM tumors in brain that is characterized by the ability to migrate and invade the adjacent healthy brain tissue. When these adherent cells were dissociated and replated at clonal density, they can regrow as sphere cultures (Fig. 3B, e) containing ~10% CD133+ cells (Fig. 3B, f).
Fig. 3.
CD133+ GBM cells can reconstitute an infiltrating GBM tumor in mouse brain that displays hypervascularity and pseudopalisading necrosis-like features. A Passaged CD133+ GBM spheres (>20 passage) can radially migrate out of spheres extensively (a–f). Magnification, 40× (f), 100× (e), 200× (a, d), 400× (b, c). B The flow cytometry analysis indicated that the adherent GBM sphere cultures contain a higher percentage of CD133+ cells (20–70%) that coexpressed SOX2 and CD44 (a–d). Replating adherent CD133+ GBM sphere culture cells at clonal cell density can re-initiate spheres that contain ~10% CD133+ GBM cells (e, f). C Genomic abnormalities that are associated with glioblastoma were detected in CD133+ GBM cells. CD133+ GBM cells were evaluated for allelic imbalances and chromosomal copy number abnormalities by using a high-density single nucleotide polymorphism array analysis. X axis, length of chromosomes 17, 10, and 7; Y axis, score of the evidence of LOH or gain of gene copy. D Intracranial injection of purified CD133+, not CD133− GBM daughter cells, can lead to the development of infiltrating tumors. HE staining shows hypercellular zones surrounding necrotic foci and the formation of a clear space (a–i). The hypervascularity was displayed by the strong positivity of CD31/PECAM-1 (platelet endothelial cell adhesion molecule-1) as determined by immunostaining (k, l). CD133 immunoreactive cells were occasionally found in small clusters (m, n). The expression of nestin, SOX2, and YKL-40 in the infiltrating cells validates the origin of human malignant GBM tumor (o–q). No immunoreactivity was determined when the control antibody was applied (j). Magnification, 200× (a–g; j–q), 400× (h, i)
CD133+ GBM cells exhibit genomic abnormalities and are capable of repopulating malignant GBM tumor in mouse brain
Both CD133+ D431 GBM cells and CD133+ S496 GBM cells exhibited loss of heterozygosity (LOH) at various chromosome locations as determined using a high-density single nucleotide polymorphism array analysis. Particularly, LOH at chromosome 10 was found in both CD133+ GBM cells, whereas only CD133+ S496 GBM cells exhibited LOH in chromosome 17 (Fig. 3C). Additionally, chromosome 7 was found to be amplified in CD133+ D431 GBM cells, and to a lesser degree in S496 cells. Similar results were found in the autologous cell line cultures passaging in the serum-containing media [28]. To compare the in vivo fate of CD133+ GBM cells and CD133− GBM cells, cells were stereotactically injected into the brains of SCID mice. Mice that received purified CD133+ GBM cells (5–10 × 103/2 μl) (11/12) sorted from the CD133+ GBM sphere cultures (6 mice per cell type) showed impaired mobility at week 15–28 post-injection, whereas mice that received CD133− GBM cells (5 × 105/2 μl) (0/20) (10 mice per cell type) remained normal at week 30. The injected CD133− GBM cells include CD133− GBM cells sorted from the same CD133+ GBM sphere cultures that were used for sorting CD133+ GBM cells (post two rounds of cell sorting) (0/6), the CD133− cells sorted from autologous GBM cell line cultured in serum (0/6) (note, serum-cultured GBM cells contain 0.2–0.35% CD133+ cells) and serum-cultured GBM cells switched to NSC culture media for 48 h (0/4) and 6 days (0/4). The H–E staining of tumors demonstrated hypercellular zones surrounding necrotic foci that form the histopathologic features of pseudopalisading necrosis as seen in human glioblastoma (Fig. 3D). Notably, S496 tumors (Fig. 3D, a–e) exhibited more enhanced necrosis than that of D431 tumors (Fig. 3D, f–i). Immunohistochemical staining revealed hypervascularity evidenced by the strong expression of CD31/platelet endothelial cell adhesion molecule-1 (PECAM-1) (Fig. 3D, k, l). The CD133 immunoreactive cells were occasionally found (Fig. 3D, m, n), suggesting the CD133− GBM daughter cells play the key role in promoting the malignant features of tumor in mice. The expression of nestin, SOX2, and YKL-40 in infiltrating tumor cells (Fig. 3D, o–q) verified the GBM origin and tumorigenic potential of CD133+ GBM cells.
Shared CD133+ GBM cell-associated genes revealed neuroectodermal properties and a quiescent/antiproliferative phenotype
To explore molecular properties of CD133+ GBM cells, we performed large-scale gene expression analysis using DNA microarrays. Based on the in vitro characterization, it is anticipated that CD133+ and CD133− daughter cells sorted from the same CD133+ sphere cultures initiated by CD133+ GBM cells will share certain properties (e.g. CD44, nestin, SOX2). Therefore, to assess genes that characterized the tumorigenic, stem-like CD133+ GBM cells, we first compared the mean level of normalized expression profiles in each of the two purified CD133+ GBM cell samples (D431 and S496) (n = 3 preparations, passage 20, 29, 40) against the non-tumorigenic, autologous CD133− GBM cells growing in serum (more differentiated condition) with and without switching to a short-term NSC culture condition (24 h, 48 h, and 6 days) (n = 6 preparations). The short-term culture of cells in NSC culture condition diminishes the likelihood of identifying NSC growth factor-responsive genes (background genes) in the comparative analysis. Probe set signals on the expression array that were ≥3-fold higher in each CD133+ GBM cell group versus the autologous CD133− GBM cell group with a pairwise t-test (P < 0.05) were selected. The filtering criteria were described in Materials and Methods. Sixty-four shared genes overexpressed in CD133+ GBM cells were identified in both pairwise comparisons (D431 and S496), and CD133/prominin1 was highly differentially expressed (D431 = 53 folds, S496 = 14 folds) as anticipated, which validates a good purification process (Table 1; Fig. 4A, a). The distinctive gene expression profiles of selected genes were verified by real-time qt-RT-PCR and semi-qt-RT-PCR (supplementary Fig. 1). The gene function enrichment analysis identified 24 significant GO clusters (Fig. 4A), and 38% (find 24 genes), 25% (find 17 genes) and 25% (find 17 genes) genes were found for GO terms related to “development” (P = 0.000001), “system development” (P = 0.000000) and “nervous system development” (P = 0.000000)), respectively (supplementary Table 1). Indeed, a large percentage of genes are neuroectoderm-developmental genes (Table 1). Most notably, Dlx5 and Dlx6 (see gene description in Table 1) are regulators of chondrogenesis of limb [33] and are highly expressed in cranial neural crest. MEOX2, MEST, and FABP4 characterized mesenchymal progenitors, and RARA [34] and Wnt antagonist FRZB [35] are likely to be inducing the signals to suppress the chondrocytic and skeletal progenitor differentiation. Simultaneously, many genes reflective of the normal function of neural crest cells were determined: SEMA6D for cardiac development, CHRNA9 for cochlea hair cell development, PPEF1 for development of cranial ganglion sensory neurons, and SHROOM2 for melanosome biogenesis. Moreover, LGR5, a novel marker gene for adult stem cells was determined [36], indicating CD133+ GBM cells share a property with other tissue stem cells.
Table 1.
Shared genes overexpressed in CD133+ D431 and CD133+ S496 GBM cells compared with autologous CD133− GBM cells cultured in serum-containing media
| Gene | Symbol | Gene I.D. | Fold change | Chromosome | Functional involvement | |
|---|---|---|---|---|---|---|
| D431 | S496 | |||||
| Sema, transmembrane, and cytoplasmic domain, 6D | SEMA6D | 80031 | 117.89 | 4.16 | 15q21 | Guidance of myocardial patterning in cardiac development |
| Growth associated protein 43 | GAP43 | 2596 | 110.32 | 25.56 | 3q13.1-q13.2 | Nervous system regeneration |
| Distal-less homeo box 6 | DLX6 | 1750 | 78.94 | 40.90 | 7q22 | Craniofacial morphogenesis/chondrogenesis |
| BH-protocadherin (brain–heart) | PCDH7 | 5099 | 62.58 | 3.76 | 4p15 | Calcium-dependent cell–cell adhesion |
| Prominin 1/CD133 | PROM1 | 8842 | 52.87 | 13.70 | 4p15.32 | Neuroepithelial stem cell marker; cell polarity |
| Endothelin 3 | EDN3 | 1908 | 43.96 | 26.35 | 20q13.2-q13.3 | Promotes neural crest cell and precursor proliferation |
| ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | ST8SIA4 | 7903 | 43.56 | 5.39 | 5q21 | Synthesis of polysialic acid in neural stem cells |
| Transcription factor AP-2 beta | TFAP2B | 7021 | 37.28 | 23.09 | 6p12 | Neural crest cell growth and differentiation |
| Distal-less homeo box 5 | DLX5 | 1749 | 36.64 | 17.18 | 7q22 | Craniofacial morphogenesis/chondrogenesis |
| Neurexin 3 | NRXN3 | 9369 | 27.84 | 5.79 | 14q31 | Stabilizes synapses |
| Cholinergic receptor, nicotinic, alpha polypeptide 9 | CHRNA9 | 55584 | 27.24 | 3.99 | 4p14 | Cochlea hair cell development |
| Fatty acid binding protein 4, adipocyte | FABP4 | 2167 | 26.43 | 70.07 | 8q21 | Lipid and glucose metabolism |
| Peptidase inhibitor 15 | PI15 | 51050 | 24.09 | 7.79 | 8q21.11 | Expressed in neuroblastoma/glioblastoma |
| Cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) | CHRNA1 | 1134 | 23.12 | 6.33 | 2q24-q32 | Neuromuscular transmission |
| Glycoprotein M6B | GPM6B | 2824 | 23.12 | 5.79 | Xp22.2 | Stabilizes proteolipids in neuron |
| Sortilin-related VPS10 domain containing receptor 1 | SORCS1 | 114815 | 19.39 | 45.42 | 10q23-q25 | Brain neuropeptide receptors |
| Protein phosphatase, EF-hand calcium binding domain 1 | PPEF1 | 5475 | 19.18 | 6.57 | Xp22.2-p22.1 | Specific sensory neuron function and/or developments |
| Kinesin family member 5C | KIF5C | 3800 | 19.03 | 17.36 | 2q23.1 | Neuronal kinesin enriched in motor neurons |
| Down syndrome critical region gene 1-like 1 | RCAN2 | 10231 | 18.71 | 10.06 | 6p12.3 | Suppresses angiogenesis |
| Leucine-rich repeat-containing G protein-coupled receptor 5 | LGR5 | 8549 | 18.56 | 4.59 | 12q22-q23 | Stem cell marker of small intestine, colon, skin, hair |
| Glutamate receptor, ionotropic, AMPA 1 | GRIA1 | 2890 | 16.48 | 14.20 | 5q31.1 | Excitatory neurotransmitter receptors |
| Mesenchyme homeo box 2 | MEOX2 | 4223 | 15.64 | 47.53 | 7p22.1-p21.3 | Somitogenesis; myogenic/sclerotomal differentiation |
| Monooxygenase, DBH-like 1 | MOXD1 | 26002 | 15.53 | 23.19 | 6q23.1-q23.3 | Dopamine-oxygenase; neural crest/ganglia marker |
| Cholinergic receptor, muscarinic 3 | CHRM3 | 1131 | 14.44 | 7.26 | 1q43 | Smooth muscle contraction; secretion of glands |
| Sulfatase 1 | SULF1 | 23213 | 13.96 | 30.43 | 8q13.2-q13.3 | Remove 6-O-sulfate groups of heparan sulfate |
| Regulator of G-protein signaling 5 | RGS5 | 8490 | 13.19 | 15.10 | 1q23.1 | Marker for pericytes; antiangiogenesis |
| Frizzled-related protein | FRZB | 2487 | 12.98 | 6.01 | 2qter | Antagonizes Wnt pathway |
| Neurocalcin delta | NCALD | 83988 | 11.78 | 3.54 | 8q22.2 | Neuronal calcium sensors; interact with S100 beta |
| Gamma-aminobutyric acid (GABA) receptor, rho 1 | GABRR1 | 2569 | 11.35 | 5.08 | 6q13-q16.3 | Reduces sensitivity to retinoic acid |
| Protocadherin 19 | PCDH19 | 57526 | 11.04 | 5.16 | Xq13.3 | Expressed in neuroepithelium |
| SRY (sex determining region Y)-box 2 | SOX2 | 6657 | 10.74 | 7.49 | 3q26.3-q27 | Neural stem cell marker, self-renewal |
| TRAF2 and NCK interacting kinase | TNIK | 23043 | 9.72 | 3.41 | 3q26.2-q26.31 | Regulates actin cytoskeleton |
| Mesoderm specific transcript homolog (mouse) | MEST | 4232 | 8.79 | 45.13 | 7q32 | Expressed in mesodermal derivatives |
| Phosphorylase kinase, gamma 2 (testis) | PHKG2 | 5261 | 8.79 | 8.01 | 16p12.1-p11.2 | Activates glycogen phosphorylase |
| Spondin 1, extracellular matrix protein | SPON1 | 10418 | 8.43 | 33.87 | 11p15.2 | Cementoblastic differentiation; inhibits angiogenesis |
| Rap guanine nucleotide exchange factor (GEF) 5 | RAPGEF5 | 9771 | 8.42 | 19.43 | 7p15.3See | RAS activator via maintain the GTP-bound state |
| Trinucleotide repeat containing 9 | TOX3 | 27324 | 8.36 | 56.65 | 16q12.1 | Regulation of neurodevelopment or neuroplasticity |
| Insulin-like growth factor binding protein 2, 36 kDa | IGFBP2 | 3485 | 7.83 | 3.09 | 2q33-q34 | Activation of the Akt and/K-Ras |
| Death-associated protein kinase 1 | DAPK1 | 1612 | 7.69 | 15.44 | 9q34.1 | Tumor suppressor |
| Formin homology 2 domain containing 3 | FHOD3 | 80206 | 7.49 | 3.15 | 18q12 | Present in nestin-expressing neuroepithelial cells |
| Scrapie responsive protein 1 | SCRG1 | 11341 | 7.09 | 80.65 | 4q31-q32 | Mesenchymal chondrogenesis, growth suppression |
| Potassium large conductance calcium-activated channel | KCNMB4 | 27345 | 7.09 | 6.00 | 12q | Smooth muscle tone and neuronal excitability |
| Retinoic acid receptor, alpha | RARA | 5914 | 6.67 | 12.21 | 17q21 | Marker for prechondrogenic progenitors |
| Neurofilament, light polypeptide 68 kDa | NEFL | 4747 | 6.6 | 6.28 | 8p21 | Controls electrical signals travel down the axon |
| Oxoglutarate (alpha-ketoglutarate) dehydrogenase | OGDH | 4967 | 6.24 | 4.36 | 7p14-p13 | Krebs cycle |
| ADAM metallopeptidase domain 23 | ADAM23 | 8745 | 5.32 | 4.38 | 2q33 | Tumor suppressor |
| Cadherin 2, type 1, N-cadherin (neuronal) | CDH2 | 1000 | 5.06 | 11.21 | 18q11.2 | Cell–cell adhesion; left-right asymmetry; cell migration |
| Immunoglobulin superfamily, member 4C | IGSF4C | 199731 | 4.75 | 10.07 | 19q13.31 | Tumor suppressor |
| Ankyrin 3, node of Ranvier (ankyrin G) | ANK3 | 288 | 4.71 | 4.19 | 10q21 | Maintenance of ion channels at nervous systems |
| Alpha-2-macroglobulin | A2M | 2 | 4.31 | 11.49 | 12p13.3-p12.3 | Protease inhibitor and cytokine transporter |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | INHBA | 3624 | 4.28 | 7.16 | 7p15-p13 | Tooth development; tumor suppressor |
| Neuropilin 2 | NRP2 | 8828 | 4.26 | 5.98 | 2q33.3 | Axon guidance in the peripheral and central neural system |
| Very low density lipoprotein receptor | VLDLR | 7436 | 4.18 | 5.83 | 9p24 | Wnt antagonist; metabolism of apoprotein-E |
| Sortilin-related VPS10 domain containing receptor 2 | SORCS2 | 57537 | 3.81 | 8.95 | 4p16.1 | Brain neuropeptide receptor |
| GalNAc-T10 | GALNT10 | 55568 | 3.67 | 4.30 | 5q33.2 | Predominant expression in CNS |
| FXYD domain containing ion transport regulator 6 | FXYD6 | 53826 | 3.49 | 22.75 | 11q23.3 | Modulator of the Na, K-ATPase |
| Leucine rich repeat neuronal 3 | LRRN3 | 54674 | 3.49 | 3.49 | 7q31.1 | Developing ganglia and motor neurons |
| Interleukin 17 receptor D | IL17RD | 54756 | 3.44 | 9.59 | 3p14.3 | Tumor suppressor-like role via FGF signaling |
| Pleiotrophin | PTN | 5764 | 3.37 | 4.86 | 7q33-q34 | Heparin binding; neurite growth-promoting factor |
| Muscleblind-like 2 (Drosophila) | MBNL2 | 10150 | 3.15 | 3.67 | 13q32.1 | Skeletal muscle development |
| Microtubule-associated protein 2 | MAP2 | 4133 | 3.09 | 4.36 | 2q34-q35 | Microtubule assembly in neurogenesis |
| Apical protein-like (Xenopus laevis) | SHROOM2 | 357 | 3.06 | 6.24 | Xp22.3 | Regulates melanosome biogenesis and localization |
| EPH receptor B3 | EPHB3 | 2049 | 3.01 | 8.71 | 3q21-qter | Precise guidance of axon and neural crest cell migration |
| FK506 binding protein 1B, 12.6 kDa | FKBP1B | 2281 | 3.00 | 4.18 | 2p23.3 | Excitation–contraction coupling in cardiac muscle |
Analysis was based on a cutoff of 3-fold increase in relative expression compared to autologous CD133− GBM cells (P < 0.05). Individual P value is shown in supplementary Table 7
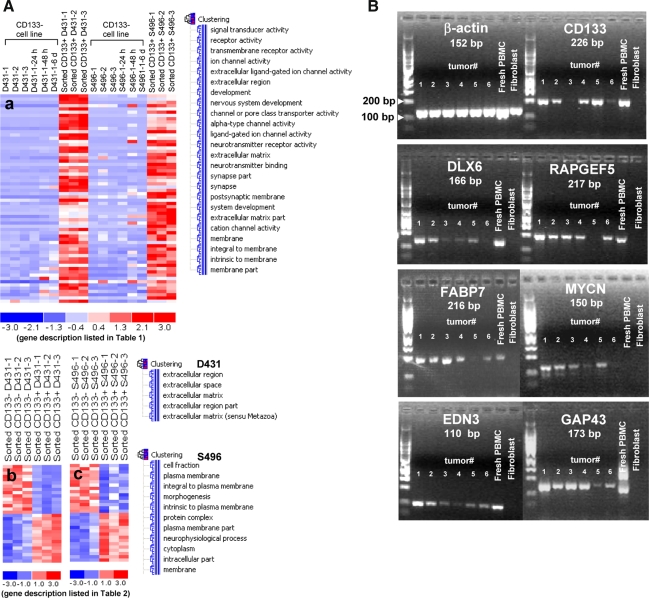
Fig. 4.
Analyses of gene expression profiles of purified, tumorigenic CD133+ GBM cells sorted from the CD133+ GBM sphere cultures. A All plots show normalized gene expression values converted into a heat map. The log2 of the fold difference is indicated by the heat map scale at the bottom. Each column is an individual sample organized into cell types and culture conditions defined at the top. Each row is a single probe set measurement of transcript abundance for an individual gene. The genes are listed in the same order from top to bottom as the corresponding tables for each of the lists. (a) All genes were filtered to select transcripts with ≥3-fold expression in the tumorigenic CD133+ GBM cells (D431 and S496) sorted from the CD133+ sphere cultures (passage (p) 20, p29, and p40) compared with the non-tumorigenic, autologous CD133− GBM cells cultured in serum-containing media (p5, p10 and p15) with or without switching to a short-term NSC culture condition for 24 h, 48 h and 6 days. Sixty-four shared genes were identified from the intersection of the comparisons between CD133+ D431 GBM cells and CD133− D431 cells, and the comparison between CD133+ S496 GBM cells and CD133− S496 GBM cells. Functional categories of gene clusters upregulated in the CD133+ GBM cells were analyzed using a gene ontology annotation–based gene function enrichment analysis (d-chip software). (b, c) Gene changes in CD133− GBM daughter cells compared to CD133+ GBM daughter cells sorted from same CD133+ GBM sphere cultures. Genes that were upregulated or down-regulated with ≥1.5-fold expression in CD133− GBM daughter cells compared with CD133+ GBM daughter cells were collected. The CD133+ and CD133− GBM cells were sorted from the sphere cultures at p20, p29, and p40. Functional categories of gene clusters in GO terms were shown. B RT-PCR analysis showed that CD133+ GBM stem cell-associated transcripts are expressed in patient-derived GBM tumors
Meanwhile, a series of genes functioning in antimitotic/antigrowth effect was identified. For instance, MEOX2 suppresses cell proliferation in a p21-dependent manner [37], SULF1 suppresses peptide growth factor signaling and angiogenesis [38], IL17RD antagonizes FGF-induced cell proliferation [39], and FRZB and VLDL act as negative regulators of the Wnt signaling pathway and angiogenesis [40]. Simultaneously, EDN3, a potent mitogen for early neural crest-derived glial and melanocytic precursors [41], and GAP-43, a crucial component of an effective neural regenerative response [42] were determined, implicating a role for maintaining the basic growth activity of CD133+ GBM cells. Thus, the overall molecular profile characterizes CD133+ GBM cells as having a slow-growing, non-differentiated, self-renewing, chondrogenic, and antidevelopmental phenotype.
Unique CD133+ GBM cell-associated genes may reflect inherently migratory properties of GBM tumor of origin
The 50 most strongly differentially expressed genes only over-expressed in each purified CD133+ GBM cells sorted from the CD133+ GBM spheres were selected (supplementary Tables 2 and 3). Uniquely, fatty acid binding protein 7 (FABP7), a migratory radial glial cell (RGC) gene, was identified as the top distinct gene in CD133+ S496. The FABP7 expressing RGC have been proposed to be the malignant glioma cell of origin [43] and the increased expression of nuclear FABP7 was found to be associated with the regions of GBM tumor infiltration, reduced survival, and recurrence [44]. On the other hand, MYCN, a migratory neural crest cell gene, and MDM2, a direct transcriptional target of MYCN, were detected in CD133+ D431. The overexpression of MDM2 is implicated in the development of de novo GBM [45]. Additional genes that are associated with cell migration machinery expressed in either CD133+ D431, CD133+ S496 or both, include genes that are associated with cell polarity (e.g. GPC3, FZD1, EPH receptor B1/B3), motor protein (KIF5C), assembly of microtubules and formation of lamellipodia and filopodia (MAP2, RHOJ, RHOU, TNIK) and formation of actin stress fibers and focal adhesions (SORBS1), pointing to an active migration characteristic of CD133+ GBM cells. More importantly, the determination of Notch effector genes (HEY1, NFIA, ID4, FABP7) reflected the prolonged Notch activation and abrogation of neurogenesis, thereby promoting a migratory phenotype and glial-fate specification [46–50]. In general, both unique gene lists are consistent with “anti-proliferative phenotype” as those of shared genes in Table 1.
CD133− GBM daughter cells divided from CD133+ GBM cells express molecular profiles associated with malignant GBM phenotype
Since CD133+ GBM cells sorted from CD133+ GBM spheres cultures expressed molecular profiles that characterized a quiescent phenotype, it is reasonable to predict that the malignant tumor-associated genes are mainly expressed in CD133− GBM daughter cells, which make up the major population of CD133+ GBM spheres and tumors. Indeed, comparative analysis of purified CD133+ and CD133− GBM daughter cells sorted from the same CD133+ GBM sphere cultures that were initiated by the purified CD133+ GBM cells revealed a transition from a tumor suppressive-like profile to tumorigenic profile when CD133+ GBM cells divided and produced CD133− GBM daughter cells. The top 15 genes in molecular changes were presented (Table 2; Fig. 4A, b and c), and the CD133 appears to be the top down-regulated gene in both pairwise comparisons. Uniquely, the gene function category analysis of gene changes in D431 cells identified 5 significant clusters all belong to extracellular component-associated GO terms (Fig. 4A, b), and 10 out of 30 genes (33%) under the GO term “extracellular region” (P value = 0.000004) (supplementary Table 4). This data thus suggests most genes that are modulated when CD133+ D431 GBM cells undergo cell division and produce fast-growing CD133− D431 daughter cells primarily include genes associated with mesenchymal/extracellular components. Evidently, two top genes, IBSP and YKL-40, determined in CD133− daughter cells are markers of osteoblast/chondrocyte differentiation and are angiogenic factors. More importantly, YKL-40 is linked to the mesenchymal and recurrent GBM phenotype [51]. Several upregulated genes further point to the early onset of inflammatory and angiogenic response. In contrast, genes that are downregulated in CD133− D431 daughter cells mostly are CD133+ D431 GBM associated genes (anti-proliferative genes) as described (Table 1; supplementary Table 2).
Table 2.
Top 15 gene changes in CD133− GBM daughter cells compared to CD133+ GBM daughter cells sorted from the same CD133+ GBM sphere cultures
| Gene | Symbol | Gene I.D. | Fold change | P value | Functional involvement |
|---|---|---|---|---|---|
| A. Genes upregulated in CD133− D431 GBM daughter cells | |||||
| Integrin-binding sialoprotein | 3381 | IBSP | 10.13 | 0.01969 | A major structural protein of the bone matrices; angiogenesis |
| Chitinase 3-like 1 (cartilage glycoprotein-39) | 1116 | CHI3L1 | 6.93 | 0.039091 | Chondrogenesis, glioblastoma progression marker |
| Tryptophan 2,3-dioxygenase | 6999 | TDO2 | 5.62 | 0.00191 | Enzyme involved in tryptophan catabolism |
| Fibronectin 1 | 2335 | FN1 | 4.61 | 0.039285 | Binds to integrins/extracellular matrix; promote tumor growth |
| Carbonic anhydrase XII | 771 | CA12 | 3.84 | 0.048487 | Acidification of the extracellular milieu; a biomarker of gliomas |
| Hydroxysteroid (11-beta) dehydrogenase 1 | 3290 | HSD11B1 | 3.75 | 0.013701 | Catalyzes the interconversion of inactive cortisone to active cortisol |
| Solute carrier family 7 | 23428 | SLC7A8 | 3.63 | 0.005935 | Transport of neutral amino acids/essential amino acids |
| Stonin 2 | 85439 | STON2 | 3.55 | 0.020396 | A component of the endocytic machinery; regulates vesicle endocytosis |
| Plexin A4, A | 57671 | PLXNA4 | 3.48 | 0.039696 | Mediates multiple semaphorin signals and regulates axon guidance |
| Calcium/calmodulin-dependent protein kinase II inhibitor 1 | 55450 | CAMK2N1 | 3.42 | 0.011008 | Inhibit brain CaM-kinase II activity |
| Anthrax toxin receptor 1 | 84168 | ANTXR1 | 3.28 | 0.031259 | Mediates cell spreading by coupling extracellular ligands to the actin |
| Growth differentiation factor 15 | 9518 | GDF15 | 3.06 | 0.004578 | Tissue differentiation and maintenance |
| Matrix metallopeptidase 14 (membrane-inserted) | 4323 | MMP14 | 3.03 | 0.01905 | Angiogenesis, tumor invasion |
| Elastin microfibril interfacer 1 | 11117 | EMILIN1 | 3.02 | 0.029463 | Extracellular matrix constituent associated with elastic fibers |
| Tissue factor pathway inhibitor | 3675 | TFPI | 3.02 | 0.000305 | Regulates the coagulation pathway; dynamic conduction of blood |
| B. Genes downregulated in CD133− D431 GBM daughter cells | |||||
| Prominin 1/CD133 | 8842 | PROM1 | −8.19 | 0.004998 | Membrane protuberances and cell polarity |
| Glutamate receptor, ionotrophic, AMPA 4 | 2893 | GRIA4 | −4.03 | 0.007852 | Excitatory neurotransmitter receptors |
| v-Myc myelocytomatosis viral related oncogene | 4613 | MYCN | −3.91 | 0.006193 | Embryonal tumor initiation factor |
| PRKC, apoptosis, WT1, regulator | 5074 | PAWR | −3.64 | 0.001191 | Tumor suppressor; apoptosis induction |
| Ksp37 protein | 83888 | KSP37 | −3.46 | 0.016226 | Protein produced by CD4 and cytotoxic lymphocytes |
| Sidekick homolog 2 (chicken) | 54549 | SDK2 | −3.43 | 0.011413 | Cell adhesion protein that guides axonal terminals |
| Frizzled-related protein | 2487 | FRZB | −3.38 | 0.00987 | Antagonizes Wnt pathway |
| Cytoplasmic FMR1 interacting protein 2 | 26999 | CYFIP2 | −3.37 | 0.002989 | A direct p53 target gene; cellular apoptosis |
| Monooxygenase, DBH-like 1 | 26002 | MOXD1 | −3.31 | 0.006728 | Predicted to hydroxylate a substrate in the endoplasmic reticulum |
| Complement factor H | 3075 | CFH | −3.13 | 0.011463 | Inhibition of complement activation |
| Activated leukocyte cell adhesion molecule | 29995 | ALCAM | −3.08 | 0.002511 | Marker of mesenchymal/colorectal cancer stem cells; growth control |
| LIM and cysteine-rich domains 1 | 214 | LMCD1 | −3.08 | 0.007018 | Represses GATA6 in the maintenance of the differentiated phenotype |
| Scrapie responsive protein 1 | 11341 | SCRG1 | −2.98 | 0.010901 | Mesenchymal chondrogenesis, growth suppression |
| v-Myb myeloblastosis viral oncogene homolog (avian) | 4602 | MYB | −2.97 | 0.049344 | Intrinsic factor for neural progenitor cell proliferation |
| Cholinergic receptor, nicotinic, alpha polypeptide 3 | 1136 | CHRNA5 | −2.97 | 0.016751 | Ligand-binding subunit of the ganglionic type nicotinic receptor |
| C. Genes upregulated in CD133− S496 GBM daughter cells | |||||
| 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 3157 | HMGCS1 | 3.09 | 0.020826 | Cholesterologenesis |
| Solute carrier family 7 | 23428 | SLC7A8 | 2.87 | 0.035311 | Transport of neutral amino acids/essential amino acids |
| Mitochondrial ribosomal protein L30 | 51263 | MRPL30 | 2.60 | 0.00943 | Protein synthesis within the mitochondrion |
| mRNA turnover 4 homolog | 51154 | MRTO4 | 2.56 | 0.007082 | mRNA turnover and ribosome assembly |
| CCAAT/enhancer binding protein zeta | 10153 | CEBPZ | 2.31 | 0.010864 | Maintains differentiated state; enhances osteoblastic differentiation |
| v-Rel reticuloendotheliosis viral oncogene homolog A | 5970 | RELA | 2.25 | 0.007044 | Cell survival, antiapoptosis |
| Endothelin 3 | 1908 | EDN3 | 2.01 | 0.04014 | Promotes neural crest cell and precursor proliferation |
| Guanine nucleotide binding protein-like 3 (nucleolar)-like | 54552 | GNL3L | 1.89 | 0.022775 | Processing of nucleolar preribosomal RNA |
| Adaptor-related protein complex 1, sigma 2 subunit | 8905 | AP1S2 | 1.89 | 0.03494 | Protein sorting and assembly of endocytic vesicles |
| Protein kinase, cAMP-dependent, catalytic, beta | 5567 | PRKACB | 1.62 | 0.04107 | Proliferation and differentiation; c-myc target gene; tumorigenesis |
| SLIT-ROBO Rho GTPase activating protein 3 | 9901 | SRGAP3 | 1.55 | 0.038237 | Negatively regulates cell migration |
| Integrin, beta 8 | 3696 | ITGB8 | 1.55 | 0.042396 | Brain vascular morphogenesis in the developing CNS |
| Monoamine oxidase A | 4128 | MAOA | 1.53 | 0.034433 | Degrades amine neurotransmitters |
| Phosphoinositide-3-kinase, class 2, beta polypeptide | 5287 | PIK3C2B | 1.53 | 0.034433 | Proliferation, survival; intracellular vesicle transport |
| Baculoviral IAP repeat-containing 4 | 331 | XIAP | 1.52 | 0.022379 | Blocks the apoptosis pathway via inhibiting caspase-3, 7, and 9 |
| D. Genes downregulated in CD133− S496 GBM daughter cells | |||||
| Prominin 1/CD133 | 8842 | PROM1 | −11.00 | 0.039802 | Membrane protuberances and cell polarity |
| Phosphorylase kinase, gamma 2 (testis) | 5261 | PHKG2 | −5.95 | 0.008141 | Activates glycogen phosphorylase |
| BH-protocadherin (brain–heart) | 5099 | PCDH7 | −2.86 | 0.013744 | Calcium-dependent cell–cell adhesion |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | 3624 | INHBA | −2.82 | 0.026005 | Tooth development; tumor suppressor |
| Melanoma cell adhesion molecule | 4162 | MCAM | −2.81 | 0.013207 | Putative adhesion molecule in neural crest cells/melanoma |
| Low density lipoprotein-related protein 1 | 4035 | LRP1 | −2.75 | 0.018718 | Lipid metabolism; antigrowth, tumor suppressor |
| Leucine rich repeat neuronal 6C | 158038 | LINGO2 | −2.69 | 0.014473 | Expressed in limbic system and neocortex |
| AF4/FMR2 family, member 3 | 3899 | AFF3 | −2.68 | 0.008324 | Regulation of lymphoid development |
| Nephronectin | 255743 | NPNT | −2.65 | 0.013665 | Tumor suppressor |
| ADAM metallopeptidase with thrombospondin type 1 motif 1 | 9510 | ADAMTS1 | −2.60 | 0.017533 | Antiangiogenesis |
| Forkhead box C1 | 2296 | FOXC1 | −2.59 | 0.022414 | Arrests cells in the G0/G1 phase; tumor suppressor |
| AT rich interactive domain 1A (SWI-like) | 8289 | ARID1A | −2.56 | 0.029044 | Differentiation-associated cell cycle arrest; tumor suppressor |
| Solute carrier family 4, anion exchanger, member 2 | 6522 | SLC4A2 | −2.52 | 0.025985 | Housekeeping regulator of intracellular pH; tumor suppressor |
| La ribonucleoprotein domain family, member 1 | 23367 | LARP1 | −2.51 | 0.01622 | Protects the 3′ end of nascent small RNAs from exonuclease digestion |
| Collagen, type IV, alpha 2 | 1284 | COL4A2 | −2.42 | 0.008793 | Inhibits angiogenesis and tumor growth |
Analysis was based on a cutoff of 1.5-fold changes in relative expression compared to CD133+ GBM daughter cells (P < 0.05)
Distinctively, most upregulated genes in sorted CD133− S496 GBM daughter cells are associated with progression of cell cycle (Table 2). The top gene, HMGCS1, is an enzyme involved in the biosynthesis of lipid/cholesterol, a critical component of biological membranes, and is thus upregulated with constant cellular proliferation [52]. Several upregulated genes further point to the early onset of proliferative differentiation, including CEBPZ, a transcription factor for maintaining the cell differentiation state, PIK3C2B, a downstream target of growth factor receptors that link to the activation of the AKT pathway, and PRKACB, a target gene of c-myc, which induces cell transformation and tumor growth [53]. As expected, down-regulated genes in sorted CD133− S496 GBM daughter cells are mostly growth regulators and tumor suppressor genes (Table 2). Indeed, 11 GO clusters that are mostly related to membrane-associated components were determined (Fig. 4A, c), and 9 (30%) and 8 (27%) genes were respectively found for GO term related to “protein complex” (P value = 0.013675) and “plasma membrane” (P = 0.020780) (supplementary Table 5). This data thus suggest when CD133+ S496 GBM cells undergo cell division and produce fast-growing CD133− S496 daughter cells primarily through intrinsic cell-cycle-based mechanism. The distinctive properties between D431 and S496 GBM cells also identified through the 2-way unsupervised gene and sample clustering of non-sorted CD133+ D431 and CD133+ S496 GBM sphere cultures, which nicely segregated two groups of samples (supplementary Fig. 2; supplementary Table 6). YKL-40 was identified as the top gene overexpressed in CD133+ D431 GBM spheres (101-fold increase) when compared to CD133+ S496 GBM spheres. By contrast, transmembrane protein 47, a gene that expressed high level transcripts in brain, was identified as the top gene overexpressed in CD133+ S496 GBM sphere cultures (233-fold increase). Correspondingly, gene function enrichment analysis showed distinct molecular pathways in the growth of two different CD133+ GBM sphere lines (mesenchymal developmental pathway versus neural developmental pathway) (supplementary Fig. 2). Uniquely, four gene clusters overexpressed in CD133+ S496 spheres were found for GO terms related to cell migration, cell adhesion, cell motility, and locomotion (supplementary Fig. 2), possibly explaining the enhanced infiltrating nature of S496 tumor in mouse brain compared to that of D431 tumor (Fig. 3D).
Expression of CD133+ GBM-associated transcripts in patients’ GBM tumors
Several CD133+ GBM-associated genes identified from the current two CD133+ GBM stem cell models have never been reported as GBM tumor-associated genes. By using RT-PCR analysis, a subset of selected CD133+ GBM-associated transcripts could be amplified in patient-derived GBM tumors (n = 6 patients) (Fig. 4B), suggesting the current culture strategy can preserve GBM stem cell properties.
Discussion
Cancer stem cell model and hypothesis has greatly changed the biological and clinical views of cancer [1–5, 54]. The molecular profiles of purified CD133+ GBM stem cells derived from the previously treated recurrent tumors characterized dormant-like cells and therefore support the hypothesis that quiescent nature of CD133+ GBM stem cells may underline the treatment resistant to the conventional therapy. The quiescent nature of cancer stem cells has been described in chronic myeloid leukemia (CML), where CML stem cells remain viable in a quiescent state even in the presence of growth factors and tyrosine kinase inhibitor [55, 56]. Indeed, while the genetic changes and tumorigenic potential were demonstrated in purified CD133+ GBM cells sorted from the CD133+ GBM spheres, the molecular profile characterized an antiproliferative nature of CD133+ GBM stem cells, suggesting the pathologic effects of molecular changes to be manifested primarily in a more differentiated progeny. Indeed, the molecular profiles of CD133+ GBM spheres (contain majority of CD133− daughter cells) initiated by the purified CD133+ GBM cells express “proliferative tumor markers” as that of CD133− GBM daughter cells, which distinctive to the quiescent CD133+ GBM daughter cells. Thus, the more differentiated CD133− GBM progeny should be considered as the true effector cells characterized fast-growing and highly angiogenic GBM tumors. The mechanisms and pathways underlying the spontaneous re-entry into active cell cycle from the quiescent state in cultures and in animal experiments remain to be elucidated [57]. In contrast to that fast-growing CD133− daughter cells, the predicted slow-cycling, non-inflammatory, and non-angiogenic nature of CD133+ GBM cells (based on the molecular profiles) may explain that GBM tumor can not be eradicated by the anti-cell cycle-based radiochemotherapy, anti-inflammatory drugs, or antiangiogenic agents.
A hallmark of all stem cells is the ability to simultaneously make identical copies of themselves (e.g. CD133+ GBM daughter cells) and give rise to a hierarchy of more differentiated progeny (e.g. CD133− GBM daughter cells). Indeed, CD133+ GBM cells fulfill this definition and are capable of undergoing cell division that give rise to a malignant tumor tissue. GBM spheres initiated with one single CD133+ GBM cell contain heterogeneous population that showed differences in cell size and proliferative potential. By RT-PCR analysis, we were able to amplify both Numb and Numb-like signals in CD133+ GBM cells (data not shown), suggesting CD133+ GBM cells may possess normal neuroepithelial-like properties capable of undergoing asymmetric cell divisions, thereby maintaining a tumor-suppressor-like phenotype [58]. Prominin/CD133 is selectively localized in protrusions of the apical membrane in neuroepithelial cells, and it was suggested that CD133 plays an important role in the maintenance of apical-basal polarity [59]. Therefore, loss of CD133 (or with other genes) may restrict CD133− GBM daughter cells to the symmetric mode of cell division and act like proliferative intermediate progenitor cells [60, 61]. Consequently, increasing the number of CD133+ GBM cells within the tumor would reflect a fast generation of proliferative and angiogenic CD133− daughter cells to form the bulk tumor [5, 62]. While our data indicated that GBM tumor growth in these two study cases depends on the CD133+ GBM cells, GBM tumor growth seemed to depend on CD133− GBM tumor-initiating cells for other cases [63, 64]. The isolation of CD133+ GBM cells from CD133− GBM cell-initiating tumor was also reported [65], indicating the CD133 is not an obligated marker for GBM stem cells.
We previously showed that GBM tumor lines established from recurrent tumors possess mesenchymal differentiation potential [29]. A recent study further showed that GBM stem cells formed tumors capable of undergoing mesenchymal differentiation [64]. The expression of mesenchymal developmental genes in CD133+ GBM stem cells may therefore provide a potential explanation for the chondrogenic/mesenchymal differentiation in tumors [66] and the shifting toward the mesenchymal phenotype upon tumor recurrence [49]. Moreover, based on the molecular profiles of purified CD133+ GBM cells, we hypothesize that the cell-of-origin of GBM tumors may be the migratory neural crest-like cell or radial glial-like cell. Overexpression of radial glial cell (RGC) marker, FABP7, in CD133+ GBM cells supports the recent finding that RGC can give rise to adult subventricular zone stem cells [67]. On the other hand, although overexpression of MYCN is associated with a childhood malignant tumor of neural crest origin, it was recently found to be one of the most frequently amplified oncogenes in GBM tumors (42%) [68]. More samples should be analyzed in order to generalize these observations. Moreover, whether these molecular properties also applied in the general properties of CD133+ GBM cells derived from the non-treated tumor remain to be investigated. Gene expression profile analyses of GBM tumors by DNA microarrays support the notion that tumor development may indeed via distinct oncogenic mechanisms among the GBM subtypes [26, 27, 69, 70]. The thought of heterogeneity in the pathway of GBM tumor development is further supported by the recent finding in studies of expression profiles of GBM sphere cultures, which showed distinct molecular properties among the GBM stem cell lines [51, 63, 64]. Likewise, although CD133+ GBM cells sorted from CD133+ D431 and CD133+ S496 spheres express shared molecular properties, unsupervised gene and sample clustering segregated two GBM sphere lines by the genes that are associated with mesenchymal developmental pathway versus neural developmental pathway, thus implying the distinct cellular-origin of these two recurrent tumors.
In summary, we characterized CD133+ GBM stem cells isolated from two tumors that are recurrent and had previous treatment. Our in vitro and in vivo data suggest that the tumorigenic CD133+ GBM stem cells are maintained at dormant-like stage state but are able to spontaneously enter the proliferative cell cycle to generate highly proliferative and angiogenic CD133− GBM daughter cells (animal data). This observation implies that tumorigenesis may be initiated through asymmetric cell division of CD133+ GBM cells [71, 72]. Thus, identifying the genes and pathways that promote the CD133+ GBM cells entering proliferative cell division cycle may facilitate the development and design of more effective therapies that specifically target the tumorigenic potential of CD133+ GBM stem cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by American Cancer Society grant #RSG-07-109-01-CCE, National Brain Tumor Foundation, The Bradley Zankel Foundation, UCLA Jonsson Cancer Center Foundation and UCLA Human Gene Medicine Seed Grant.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11060-009-9919-z) contains supplementary material, which is available to authorized users.
References
- 1.Galli R, Binda E, Orfanelli U (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021. doi:10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed]
- 2.Hemmati HD, Nakano I, Lazareff JA et al (2003) Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA 100:15178–15183. doi:10.1073/pnas.2036535100 [DOI] [PMC free article] [PubMed]
- 3.Singh SK, Hawkins C, Clarke ID et al (2004) Identification of human brain tumour initiating cells. Nature 432:396–401. doi:10.1038/nature03128 [DOI] [PubMed]
- 4.Yuan X, Curtin J, Xiong Y et al (2004) Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23:9392–9400. doi:10.1038/sj.onc.1208311 [DOI] [PubMed]
- 5.Bao S, Wu Q, McLendon RE et al (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760. doi:10.1038/nature05236 [DOI] [PubMed]
- 6.Loeffler M, Roeder I (2002) Tissue stem cells: definition, plasticity, heterogeneity, self-organization and models—a conceptual approach. Cells Tissues Organs 171:8–26. doi:10.1159/000057688 [DOI] [PubMed]
- 7.Fuchs E, Segre JA (2000) Stem cells: a new lease on life. Cell 100:143–155. doi:10.1016/S0092-8674(00)81691-8 [DOI] [PubMed]
- 8.Lavker RM, Sun TT (2000) Epidermal stem cells: properties, markers, and location. Proc Natl Acad Sci USA 97:13473–13475. doi:10.1073/pnas.250380097 [DOI] [PMC free article] [PubMed]
- 9.Perryman SV, Sylvester KG (2006) Repair and regeneration: opportunities for carcinogenesis from tissue stem cells. J Cell Mol Med 10:292–308. doi:10.1111/j.1582-4934.2006.tb00400.x [DOI] [PMC free article] [PubMed]
- 10.Fuchs E, Tumbar T, Guasch G (2004) Socializing with the neighbors: stem cells and their niche. Cell 116:769–778. doi:10.1016/S0092-8674(04)00255-7 (Review) [DOI] [PubMed]
- 11.Moore KA, Lemischka IR (2006) Stem cells and their niches. Science 311:1880–1885. doi:10.1126/science.1110542 [DOI] [PubMed]
- 12.Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL (1997) Identification of a lineage of multipotent hematopoietic progenitors. Development 124:1929–1939 [DOI] [PubMed]
- 13.Schmidt GH, Winton DJ, Ponder BA (1988) Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development 103:785–790 [DOI] [PubMed]
- 14.Oshima H, Rochat A, Kedzia C (2001) Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104:233–245. doi:10.1016/S0092-8674(01)00208-2 [DOI] [PubMed]
- 15.Jensen UB, Lowell S, Watt FM (1999) The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on the whole mount labelling and lineage analysis. Development 126:2409–2418 [DOI] [PubMed]
- 16.Jones PH, Watt FM (1993) Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73:713–724. doi:10.1016/0092-8674(93)90251-K [DOI] [PubMed]
- 17.Hong KU, Reynolds SD, Giangreco A et al (2001) Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 24:671–681 [DOI] [PubMed]
- 18.Borthwick DW, Shahbazian M, Krantz QT et al (2001) Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol 24:662–670 [DOI] [PubMed]
- 19.Reynolds SD, Giangreco A, Hong KU et al (2004) Airway injury in the pathophysiology of lung disease: selective depletion of airway stem and progenitor cells pools potentates lung inflammation and alveolar dysfunction. Am J Physiol Lung Cell Mol Physiol 287:L1256–L1265. doi:10.1152/ajplung.00203.2004 [DOI] [PubMed]
- 20.Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3:730–737. doi:10.1038/nm0797-730 [DOI] [PubMed]
- 21.Vermeulen L, Todaro M, de Sousa Mello F et al (2008) Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA 105:13427–13432. doi:10.1073/pnas.0805706105 [DOI] [PMC free article] [PubMed]
- 22.Dalerba P, Cho RW, Clarke MF (2007) Cancer stem cells: models and concepts. Annu Rev Med 58:267–284. doi:10.1146/annurev.med.58.062105.204854 (Review) [DOI] [PubMed]
- 23.Beachy PA, Karhadkar SS, Berman DM (2004) Tissue repair and stem cell renewal in carcinogenesis. Nature 432:324–331. doi:10.1038/nature03100 [DOI] [PubMed]
- 24.Zhang QB, Ji XY, Huang Q et al (2006) Differentiation profile of brain tumor stem cells: a comparative study with neural stem cells. Cell Res 16:909–915. doi:10.1038/sj.cr.7310104 [DOI] [PubMed]
- 25.Clarke MF, Dick JE, Dirks PB et al (2006) Cancer stem cells—perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res 66:9339–9344 [DOI] [PubMed]
- 26.Freije WA, Castro-Vargas FE, Fang Z et al (2004) Gene expression profiling of gliomas strongly predicts survival. Cancer Res 64:6503–6510. doi:10.1158/0008-5472.CAN-04-0452 [DOI] [PubMed]
- 27.Tso CL, Freije WA, Day A et al (2006) Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res 66:159–167. doi:10.1158/0008-5472.CAN-05-0077 [DOI] [PubMed]
- 28.Tso CL, Shintaku P, Chen J et al (2006) Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res 4:607–619. doi:10.1158/1541-7786.MCR-06-0005 [DOI] [PubMed]
- 29.Guan Y, Gerhard B, Hogge DE (2003) Detection, isolation, and stimulation of quiescent primitive leukemic progenitor cells from patients with acute myeloid leukemia (AML). Blood 101:3142–3149. doi:10.1182/blood-2002-10-3062 [DOI] [PubMed]
- 30.Bez A, Corsini E, Curti D et al (2003) Neurosphere and neurosphere-forming cells: morphological and ultrastructural characterization. Brain Res 993:18–29. doi:10.1016/j.brainres.2003.08.061 [DOI] [PubMed]
- 31.Flanagan LA, Rebaza LM, Derzic S et al (2006) Regulation of human neural precursor cells by laminin and integrins. J Neurosci Res 83:845–856. doi:10.1002/jnr.20778 [DOI] [PMC free article] [PubMed]
- 32.Jacques TS, Relvas JB, Nishimura S et al (1998) Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development 125:3167–3177 [DOI] [PubMed]
- 33.Hsu SH, Noamani B, Abernethy DE et al (2006) Dlx5- and Dlx6-mediated chondrogenesis: differential domain requirements for a conserved function. Mech Dev 123:819–830. doi:10.1016/j.mod.2006.08.005 [DOI] [PubMed]
- 34.Weston AD, Rosen V, Chandraratna RA et al (2000) Regulation of skeletal progenitor differentiation by the BMP and retinoid signaling pathways. J Cell Biol 148:679–690. doi:10.1083/jcb.148.4.679 [DOI] [PMC free article] [PubMed]
- 35.Enomoto-Iwamoto M, Kitagaki J, Koyama E et al (2002) The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol 251:142–156. doi:10.1006/dbio.2002.0802 [DOI] [PubMed]
- 36.Barker N, van Es JH, Kuipers J et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007. doi:10.1038/nature06196 [DOI] [PubMed]
- 37.Chen Y, Leal AD, Patel S (2007) The homeobox gene GAX activates p21WAF1/CIP1 expression in vascular endothelial cells through direct interaction with upstream AT-rich sequences. J Biol Chem 282:507–517. doi:10.1074/jbc.M606604200 [DOI] [PMC free article] [PubMed]
- 38.Narita K, Staub J, Chien J et al (2006) HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res 66:6025–6032. doi:10.1158/0008-5472.CAN-05-3582 [DOI] [PubMed]
- 39.Kovalenko D, Yang X, Nadeau RJ et al (2003) Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J Biol Chem 278:14087–14091. doi:10.1074/jbc.C200606200 [DOI] [PubMed]
- 40.Chen Y, Hu Y, Lu K et al (2007) Very low density lipoprotein receptor, a negative regulator of the wnt signaling pathway and choroidal neovascularization. J Biol Chem 282:34420–34428. doi:10.1074/jbc.M611289200 [DOI] [PubMed]
- 41.Lahav R, Dupin E, Lecoin L (1998) Endothelin 3 selectively promotes survival and proliferation of neural crest-derived glial and melanocytic precursors in vitro. Proc Natl Acad Sci USA 95:14214–14219. doi:10.1073/pnas.95.24.14214 [DOI] [PMC free article] [PubMed]
- 42.Shen Y, Mani S, Meiri KF (2004) Failure to express GAP-43 leads to disruption of a multipotent precursor and inhibits astrocyte differentiation. Mol Cell Neurosci 26:390–405. doi:10.1016/j.mcn.2004.03.004 [DOI] [PubMed]
- 43.Mita R, Coles JE, Glubrecht DD et al (2007) B-FABP-expressing radial glial cells: the malignant glioma cell of origin? Neoplasia 9:734–744. doi:10.1593/neo.07439 [DOI] [PMC free article] [PubMed]
- 44.Kaloshi G, Mokhtari K, Carpentier C et al (2007) FABP7 expression in glioblastomas: relation to prognosis, invasion and EGFR status. J Neurooncol 84:245–248. doi:10.1007/s11060-007-9377-4 [DOI] [PubMed]
- 45.Biernat W, Kleihues P, Yonekawa Y et al (1997) Amplification and overexpression of MDM2 in primary (de novo) glioblastomas. J Neuropathol Exp Neurol 56:180–185. doi:10.1097/00005072-199702000-00009 [DOI] [PubMed]
- 46.Edenfeld G, Altenhein B, Zierau A et al (2007) Notch and Numb are required for normal migration of peripheral glia in Drosophila. Dev Biol 301:27–37. doi:10.1016/j.ydbio.2006.11.013 [DOI] [PubMed]
- 47.Sakamoto M, Hirata H, Ohtsuka T et al (2003) The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem 278:44808–44815. doi:10.1074/jbc.M300448200 [DOI] [PubMed]
- 48.Deneen B, Ho R, Lukaszewicz A et al (2006) The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52:953–968. doi:10.1016/j.neuron.2006.11.019 [DOI] [PubMed]
- 49.Jeon HM, Jin X, Lee JS et al (2008) Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev 22:2028–2033. doi:10.1101/gad.1668708 [DOI] [PMC free article] [PubMed]
- 50.Anthony TE, Mason HA, Gridley T et al (2005) Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev 19:1028–1033. doi:10.1101/gad.1302105 [DOI] [PMC free article] [PubMed]
- 51.Phillips HS, Kharbanda S, Chen R et al (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173. doi:10.1016/j.ccr.2006.02.019 [DOI] [PubMed]
- 52.Coleman PS (1986) Membrane cholesterol and tumor bioenergetics. Ann N Y Acad Sci 488:451–467. doi:10.1111/j.1749-6632.1986.tb46578.x [DOI] [PubMed]
- 53.Wu KJ, Mattioli M, Morse HC et al (2002) c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cbeta) gene. Oncogene 21:7872–7882. doi:10.1038/sj.onc.1205986 [DOI] [PubMed]
- 54.Al-Hajj M, Becker MW, Wicha M et al (2004) Therapeutic implications of cancer stem cells. Curr Opin Genet Dev 14:43–47. doi:10.1016/j.gde.2003.11.007 (Review) [DOI] [PubMed]
- 55.Holyoake T, Jiang X, Eaves C, Eaves A (1999) Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood 94:2056–2064 [PubMed]
- 56.Graham SM, Jørgensen HG, Allan E et al (2002) Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99:319–325. doi:10.1182/blood.V99.1.319 [DOI] [PubMed]
- 57.Sang L, Coller HA, Roberts JM (2008) Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321:1095–1100. doi:10.1126/science.1155998 [DOI] [PMC free article] [PubMed]
- 58.Chia W, Somers WG, Wang H (2008) Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol 180:267–272. doi:10.1083/jcb.200708159 [DOI] [PMC free article] [PubMed]
- 59.Kosodo Y, Roper K, Haubensak W et al (2004) Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J 23:2314–2324. doi:10.1038/sj.emboj.7600223 [DOI] [PMC free article] [PubMed]
- 60.Pontious A, Kowalczyk T, Englund C et al (2008) Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci 30:24–32. doi:10.1159/000109848 [DOI] [PubMed]
- 61.Bello BC, Izergina N, Caussinus E et al (2008) Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev 3:5. doi:10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed]
- 62.Zeppernick F, Ahmadi R, Campos B et al (2008) Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res 14:123–129. doi:10.1158/1078-0432.CCR-07-0932 [DOI] [PubMed]
- 63.Gunther HS, Schmidt NO, Phillips HS et al (2008) Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 27:2897–2909. doi:10.1038/sj.onc.1210949 [DOI] [PubMed]
- 64.Beier D, Hau P, Proescholdt M et al (2007) CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res 67:4010–4015. doi:10.1158/0008-5472.CAN-06-4180 [DOI] [PubMed]
- 65.Wang J, Sakariassen PØ, Tsinkalovsky O et al (2008) CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer 122:761–768. doi:10.1002/ijc.23130 [DOI] [PubMed]
- 66.Ricci-Vitiani L, Pallini R, Larocca LM et al (2008) Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ 15:1491–1498. doi:10.1038/cdd.2008.72 [DOI] [PubMed]
- 67.Merkle FT, Tramontin AD, Garcia-Verdugo JM et al (2004) Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA 101:17528–17532. doi:10.1073/pnas.0407893101 [DOI] [PMC free article] [PubMed]
- 68.Hui AB, Lo KW, Yin XL et al (2001) Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest 81:717–723 [DOI] [PubMed]
- 69.Nutt CL, Mani DR, Betensky RA et al (2003) Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res 63:1602–1607 [PubMed]
- 70.Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170(5):1445–1453. doi:10.2353/ajpath.2007.070011 Review [DOI] [PMC free article] [PubMed]
- 71.Lee M, Vasioukhin V (2008) Cell polarity and cancer-cell and tissue polarity as a non-canonical tumor suppressor. J Cell Sci 121:1141–1150. doi:10.1242/jcs.016634 [DOI] [PubMed]
- 72.Wodarz A, Nathke I (2007) Cell polarity in development and cancer. Nat Cell Biol 9:1016–1024. doi:10.1038/ncb433 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.