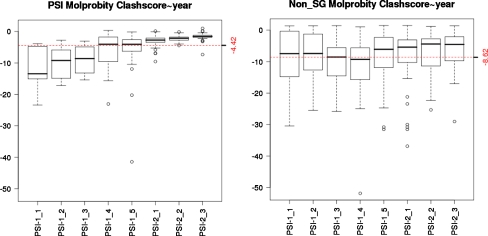
Fig. 2.
NMR structure quality assessed by MolProbity [7], using Z scores defined in Protein Structure Validation Suite (PSVS) software [5], are significantly better for structures generated in the PSI Centers (median Z score = −4.4 over 8 years) compared with NMR structures deposited in the PDB by groups not involved in structural genomics (median Z score = −8.6), during the same time periods. The analysis included statistics for all NMR structures deposited in the PDB by PSI centers (left panel) compared with statistics for 50 NMR structures chosen randomly from those deposited in the PDB by nonSG research groups (right panel) during the same time period. In these box plots, the central horizontal line is the median value, the bottom and top of the box denote the first and third quantiles, and the vertical “whiskers” denote ±1.5 times the interquartile range (approximately two standard deviations). Outlier values are indicated by open circles