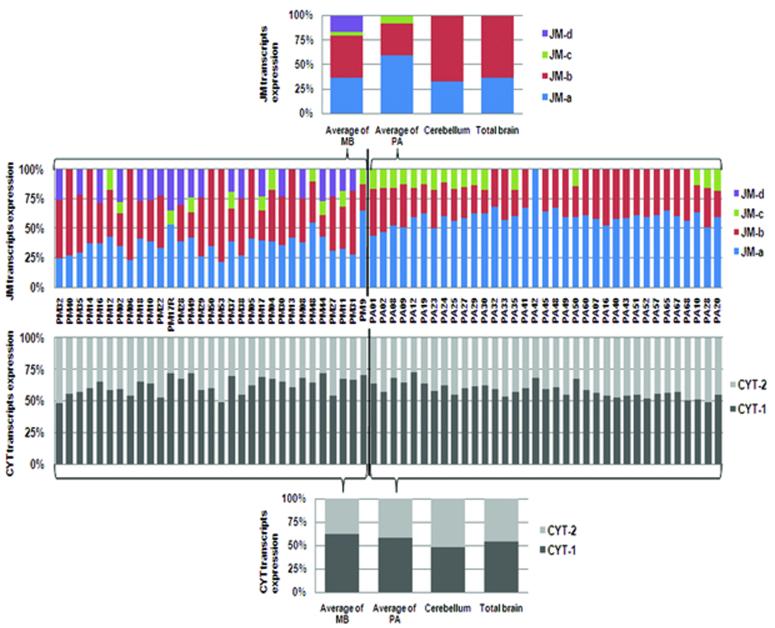
FIGURE 4.
Composition of ERBB4 transcript variants in individual tumour samples and normal brain. LightCycler Hybridization Probes were used to specifically detect the junctions between alternatively spliced exons representing the ERBB4 transcript variants JM-a, JM-b, JM-c, JM-d, CYT-1 and CYT-2. The composition of ERBB4 variants in individual tumours was calculated as the percentage of the total JM or CYT expression of each transcript variant. The upper panel shows the composition of JM-region transcript variants and the lower panel shows the CYT variants. The average expression pattern of 31 MB tumours and 35 PA tumours was also calculated and used in comparison with normal brain tissues (cerebellum and total brain). SDs of two parallel analyses were < 5% of the means. MB, medulloblastoma; PA, pilocytic astrocytoma.