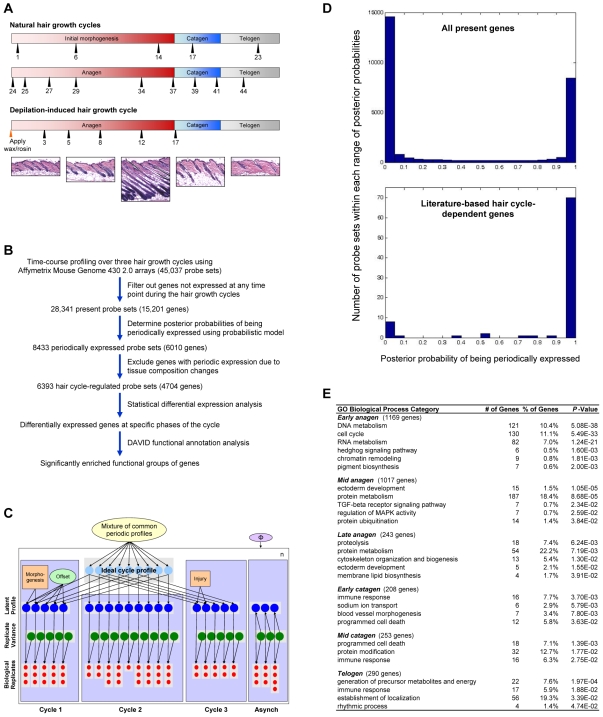
Figure 1. Identification of hair cycle–regulated genes using probabilistic models.
(A) Representative time points selected for gene expression profiling of the natural and depilation-induced hair growth cycles. Time points for natural cycles represent postnatal days, and time points for depilation-induced cycle indicate the number of days following depilation. Since the three cycles have different duration, the schematic timeline is not in actual time scale. Bottom panel consists of representative histology of dorsal skin at key phases of synchronized hair follicle cycling. (B) Overview of data processing and statistical analyses. Note that samples for the first synchronized hair growth cycle and asynchronous time points were profiled in an earlier study using an older generation array (Affymetrix Murine Genome U74Av2). (C) Schematic of the probabilistic model for detection of periodic gene expression changes during hair follicle cycling. See Materials and Methods for a detailed description of the schematic. Asynch – asynchronous cycle. (D) Histogram of the number of probe sets within each range of the indicated posterior probabilities of being periodically expressed. Top panel is the histogram of probabilities for all present genes during the hair growth cycle. The bottom panel is the histogram of probabilities for the literature-based hair cycle-dependent genes. (E) List of significantly enriched GO Biological Process categories within the sets of genes specifically upregulated at the indicated phases of the hair growth cycle. Due to the redundancy of categories, not all are listed. The number of genes upregulated at each phase of the cycle is in parentheses. Statistical enrichment of each category is shown as P-values calculated using a modified Fisher Exact test (DAVID functional annotation analysis).