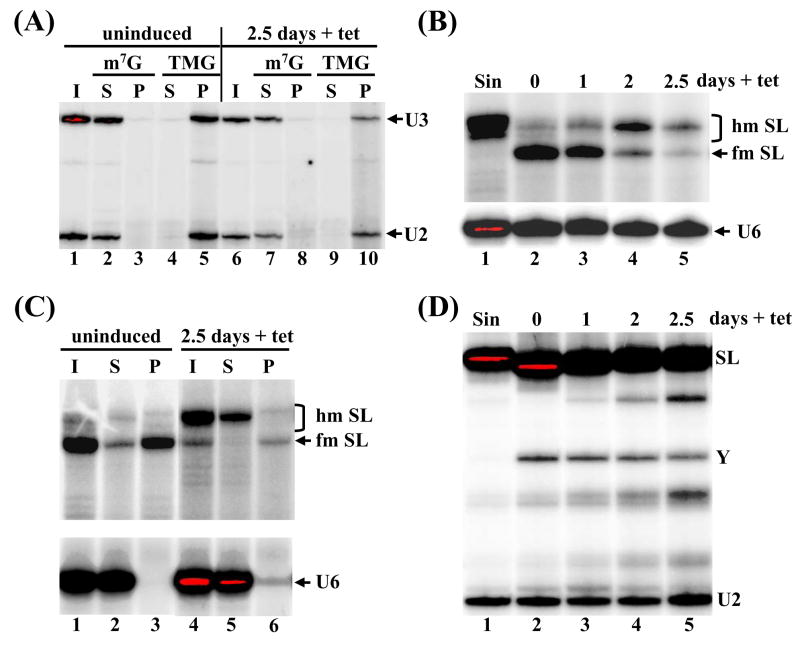
Fig. 4.
TbCGM1 depletion affects SL RNA capping. (A) Total RNA isolated from uninduced (lanes 1–5) or TbGM1-RNAi cells induced for 2.5 days (lanes 6–10) was immunoprecipitated with anti-m7G and anti-TMG antibodies and the input (I), supernatant (S) and pellet (P) fractions were assayed by primer extension of the U2 snRNA (U2) and U3 snoRNA (U3). (B) The TbCGM1 RNAi cell line was induced with tet for the indicated number of days (lanes 2–5) and total RNA was assayed by primer extension with an SL intron-specific primer. The position of fully-modified (fm SL) and hypomodified (hm SL) SL RNA is indicated. RNA isolated from sinefungin-treated cells served as a control for hypomodified SL RNA (lane 1). A U6 snRNA-specific primer was included in the reactions to control for RNA amounts and quality. (C) Total RNA isolated from uninduced cells (lanes 1–3) or from cells depleted of TbCgm1 for 2.5 days (lanes 4–6) was immunoprecipitated with anti-m7G antibodies and the input (I), supernatant (S) and pellet (P) fractions were assayed by primer extension with an SL intron-specific primer. A U6-snRNA-specific primer was included in all the reactions to control for immunoprecipitation specificity (U6). (D) Primer extension of total RNA with an oligonucleotide complementary to nt 110 to 131 of the SL RNA. The position of the Y-structure intermediate (Y) and full-length SL RNA (SL) is indicated. RNA isolated from sinefungin-treated cells was used as a control for trans-splicing inhibition (lane 1). An oligonucleotide complementary to the U2 snRNA was included as a loading and quality control (U2).