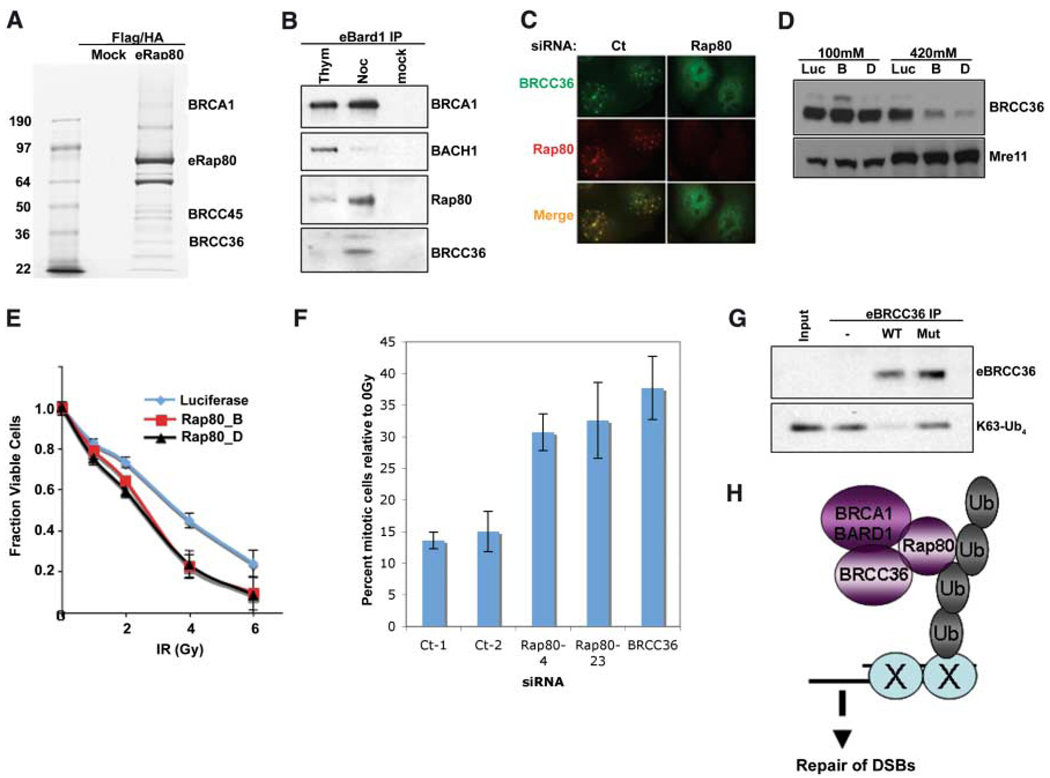
Fig. 4.
Interactions of RAP80, BRCC36, and BRCA1. (A) Coommassiestained gel of FLAG and HA double-purified RAP80 complexes from HeLa S3 cells stably expressing eRAP80. (B) HeLa S3 cells stably expressing eBARD1 were treated with 2 mM thymidine (Thym) or 0.5 µM nocodazole (Noc). FLAG-purified proteins were analyzed by IB. (C) U2OS cells expressing HA-tagged BRCC36 were transfected with shRNA specific to RAP80 or control and, 48 hours later, were monitored for IRIF at 1 hour after 6 Gy. (D) HeLa cells expressing stable siRNA vectors specific to luciferase (Luc) or RAP80 (B and D) were sequentially extracted with NETN buffer containing 100 mM NaCl and then NETN buffer containing 420 mM NaCl. Extracts were analyzed by IB. (E) HeLa cells expressing stable shRNA vectors specific to luciferase or RAP80 (B and D) were treated with the indicated doses of IR, and the number of viable cells were counted 96 hours later. The fraction of viable cells compared with a culture receiving 0 Gy is expressed graphically. Experiments were done in triplicate. Error bars indicate standard deviation. (F) The percentage of phosphohistone H3– positive cells (mitotic population) at 1 hour after 2 Gy IR compared with at 0 Gy (i.e., mock irradiation) was plotted for each siRNA-transfected U2OS population. Experiments were done in triplicate; error bars indicate standard deviation. (G) FLAG peptide eluates from FLAG IPs performed on extracts of HeLa S3 cells stably expressing FLAG-HA–tagged WT or a catalytically inactive BRCC36 mutant were incubated with K63-linked tetraubiquitin (K63-Ub4) for 30 min at 37°C. The reaction mixtures were then analyzed by IB. (H) Model for recruitment of a BRCA1-BARD1-BRCC36-RAP80 complex to sites of DNA damage by binding to non-K48–linked ubiquitin structures.