Abstract
Multiple Reaction Monitoring (MRM), commonly employed for the mass spectrometric detection of small molecules, is rapidly gaining ground in proteomics. Its high sensitivity and specificity makes this targeted approach particularly useful when sample throughput or proteome coverage limits global studies. Existing tools to design MRM assays rely exclusively on theoretical predictions, or combine them with previous observations on the same type of sample. The additional mass spectrometric experimentation this requires can pose significant demands on time and material. To overcome these challenges, a new MRM worksheet was introduced into The Global Proteome Machine database (GPMDB) that provided all of the information needed to design MRM transitions based solely on archived observations made by other researchers in previous experiments. This required replacing the precursor ion intensity by the number of peptide observations, which proved to be an adequate substitute if peptides did not occur in multiple forms. While the absence of collision energy information proved largely inconsequential, successful prediction of unique transitions depended on the type of fragment ion involved. The design of MRM assays for iTRAQ-labeled tryptic peptides obtained from human platelet proteins demonstrated the usefulness of the MRM worksheet also for quantitative applications. This workflow, which relies exclusively on experimental observations stored in data repositories, therefore represents an attractive alternative for the prediction of MRM transitions prior to experimental validation and optimization.
Keywords: Tandem Mass Spectrometry, Multiple Reaction Monitoring, Targeted Proteomics, Data Repository, Platelets
Introduction
The global, unbiased analysis of a proteome is an important aspect of systems biology, as proteins are involved in the majority of processes taking place in a biological system [1]. Mass spectrometry allows the characterization of large numbers of proteins without prior knowledge of their identity, making it the method of choice in global proteome analyses [2]. Despite significant technical advances, current mass spectrometers are still not capable of handling the complexity and dynamic range of the peptide mixtures that are generated in proteomics. As a result, such mixtures are typically undersampled, i.e. not all of the peptides present in the mixture are isolated and subjected to fragmentation to produce sequence information. This mostly affects peptides of lower intensity that co-elute with more abundant peptides, as they will escape further analysis in the standard experimental setup that selects the peptides to be sequenced based on signal intensity [3].
Several options exist to overcome this limitation: iterative analyses that exclude already sequenced peptides can be carried out [4]; peptides of particular interest can be given priority over others based on inclusion lists [5]; or dedicated scans can be performed that focus on a subset of peptides carrying specific information, for example precursor ion scans to detect post-translationally modified peptides such as phosphopeptides [6]. The latter two approaches are targeted, as they take advantage of prior knowledge of a peptide’s mass-to-charge ratio (m/z) or a characteristic fragment ion, respectively, to increase the specificity and sensitivity of its detection. Extending this concept further, i.e. defining the m/z of the peptide and one of its fragment ions, results in the maximum specificity and sensitivity possible for mass spectrometric peptide detection. Indeed, the corresponding Multiple Reaction Monitoring (MRM) approach is commonly applied in pharmaceutical research for the identification of trace amounts of analyte molecules in complex matrices [7, 8]. In proteomics, an increasing number of reports demonstrate its suitability for the detection of low abundant proteins in plasma [9, 10] and post-translational modification sites in proteins [11–15], and for quantitative comparisons between different proteome states [16–18].
As a targeted proteomics approach, MRM requires the definition of the experimental parameters prior to sample analysis. For the highly sensitive analysis of proteins, this translates into knowing the m/z of an abundant, consistently produced (or “proteotypic”) peptide [19], as well as the m/z of one of its fragment ions that is generated with high intensity. Two different general workflows to predict the most promising set of suitable MRM transitions have been realized to date: while one approach relies entirely on physicochemical parameters and in silico predictions [9, 19], a recently introduced alternative expands this concept by also taking into account archived experimental information obtained in previous global proteome experiments [20]. Both approaches involve substantial additional experimentation prior to MRM assay validation and optimization, as they require the evaluation of the predicted sets of MRM transitions or the initial definition of the proteome under study. In contrast, a workflow that relies exclusively on previous experimental observations stored in data repositories to design MRM assays would potentially reduce the amount of time and material consumed in these initial steps.
Here, we describe the utilization of The Global Proteome Machine database (http://gpmdb.thegpm.org), currently the largest curated repository for tandem mass spectrometric data, for this purpose. To this end, a novel MRM worksheet was developed and implemented that provides the necessary information for the design of MRM assays. Using tryptic digests of human platelet lysates, this interface was then tested for its suitability and usefulness for this application. Examination of the information available in GPMDB highlighted the lack of two parameters linked to the sensitivity of an MRM transition: precursor ion intensity and collision energy. Subsequent investigation showed the number of peptide observations to be an adequate substitute for precursor ion intensity, whereas the lack of information on the collision energy was found to be largely inconsequential for the success rate of the initial tests. Utilization of GPMDB for the prediction of the uniqueness of a transition had limited success. In contrast, the design of suitable MRM transitions for iTRAQ-labeled peptides required only minimal manipulation that affected the sensitivity of individual transitions, but not the specificity or general usefulness. Taken together, these results demonstrate that the new MRM worksheet in GPMDB can be used successfully for the initial design of MRM transitions that monitor tryptic peptides from human proteins in complex samples.
Materials and Methods
MRM Library Creation, Storage and Retrieval
The MRM spectrum library used in this study was derived from data gathered by the Global Proteome Machine project since its inception in January 2003. Approximately 600,000,000 peptide tandem mass spectra generated by many laboratories using a variety of experimental protocols and analytical instrumentation were analyzed using one of the X! Series search engines (X! Tandem [21], PPP [22] or Hunter [23]) and 5.9 × 107 of the highest quality peptide sequence assignments, along with the information required for the statistical analysis of the sequence assignment and the corresponding tandem mass spectrum were stored in data repository that consists of a collection of XML analysis files and a database, collectively referred to as GPMDB [24]. On average, each peptide sequence was represented by 46 separate observations.
The information in GPMDB on human source proteins was used to generate a set of consensus spectra representing all of the peptide sequence assignments with an expectation of being a stochastic assignment (E-value) of less than 0.001. The set of spectra associated with a unique peptide sequence were clustered into multiple subsets. These subsets were based on the precursor ion charge and spectral characteristics of the individual tandem mass spectra and the observed modifications of the peptide sequence: post-translational modifications; experimental artifacts and reagents (e.g., methionine oxidation or cysteine blocking reagents); and reagents for quantitation using isotope dilution methods. These subsets were combined into consensus spectra using a method previously described [23]. Each unique peptide sequence/modification state/precursor ion charge combination could result in multiple consensus spectra, based on the observed pattern of fragment ions. These additional subsets were primarily based on whether low mass (< 300 Da) ions were observed or not. This pattern-based clustering was designed to differentiate between tandem spectra collected on mass spectrometers with ion trap mass analyzers and those collected with quadrupole-TOF analyzers for doubly and triply charged precursor ions. Spectra from singly charged precursor ions were dominated by data from TOF-TOF analyzers with MALDI ion sources.
The resulting consensus spectra were then subjected to a further set of curation steps, prior to inclusion in the final MRM library. To be included, a spectrum must have had at least 60 % of the total number of ions assigned as b- or y-ions or their corresponding secondary fragment ions after neutral loss of ammonia or water, and these ions must have contributed at least 60 % of its total intensity. The m/z value for these assignable ions were corrected to be the true calculated values, in the final library. Any spectrum associated with either an isotopically labeled peptide, a quantitation reagent (such as iTRAQ or ICAT) or a single nucleotide-induced amino acid polymorphism (SNAP) was excluded from the library. The peptide sequences were then aligned with the protein sequences in the most recent version of the human genome assembly (NCBI 36) using the ENSEMBL gene models (Homo sapiens, version 50) and the protein sequence coordinates recorded.
Once the libraries had been created and curated, they were loaded into a relational database, constructed using MySQL (version 5.0). The user interface was constructed to be readable in any commonly used HTML web browser, using the Apache HTTP server (version 2.2) and Perl (version 5.0). All of the displayed pages were generated from the database contents at runtime, with no stored pages or procedures. The source code used to load the libraries, construct the database and render the various displays was made available at the GPM project FTP site (ftp://ftp.thegpm.org/repos/mrm). The peptide frequency information used in the displays was generated from the GPMDB daily built and the relative retention time information was calculated using a previously described method [25].
Preparation of platelets
Ethical approval for this study was granted by the University of British Columbia Research Ethics Board and informed consent was granted by the donors. Whole blood was drawn from the antecubital vein of healthy human volunteers into 0.15 % (v/v) acid-citrate-dextrose (ACD) anticoagulant. Standard centrifugation procedures were used to isolate platelets, which were then washed twice in physiological buffer (10 mM trisodium citrate, 30 mM dextrose, 1 U/mL apyrase). Platelets were re-suspended in Tris-buffered saline containing 5 mM EDTA, and counted using an Advia Haematology Analyzer. Platelets were centrifuged at 1500 rpm for 15 minutes at room temperature and the platelet pellet was resuspended in 1× lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 % TX-100, 2.5 mM Na pyrophosphate, 1 mM β-glycerolphosphate, 1 mM Na3VO4, 1 × protease inhibitor cocktail). Platelets were lysed for an hour on ice prior to storage at −80°C if required.
Proteolytic Digestion
Platelet lysate samples, containing 250 µg protein, were thawed slowly on ice and exchanged into 25 µl of 50 mM NH4HCO3 by centrifugation using 3000 Da Molecular Weight Cutoff Microcon centrifugal filter devices (Millipore Corporation, Bedford, MA, USA). Samples were denatured by heating at 99°C for 10 minutes, reduced using 1 µg dithiothreitol (DTT) per 50 µg protein (5 µL of 1 mg/mL DTT stock solution) at 37°C for 30 minutes and alkylated with 5 µg iodoacetamide per 50 µg protein (25 µL of 1mg/mL iodoacetamide stock solution) at 37°C for 20 minutes. In solution tryptic digestions were performed overnight at 37°C, using 1 µg trypsin per 50 µg protein (5 µL of 1 mg/mL trypsin stock solution). Digestion was stopped by addition of 1 µL concentrated Trifluoracetic acid (TFA).
Sample separation and clean-up
Strong cation exchange (SCX) was conducted using an Ettan MDLC (GE Healthcare Biosciences AB, Uppsala, Sweden). The column used was a 4.6 mm × 150 mm Zorbax 300-SCX column with particle size 5 µm and sulfonic acid-based cation exchange ligand (Agilent Technologies, Santa Clara, CA, USA). The mobile phases for running the SCX separation were 25 % HPLC grade acetonitrile (ACN, buffer A) and 25 % HPLC grade ACN and 500 mM NH4Cl (buffer B). The gradient for Buffer B changed from 0 − 60 % over 20 minutes, and from 60 % − 100 % over five minutes using a flow rate of 30 µL/min. Twenty-five fractions (each 500 µL) were collected. Fractions were dried down and re-suspended in 20 µL 5 % formic acid (FA). All of the fractions were Zip-Tipped individually using OMIX C18 tips (Varian Inc, Lake Forest, CA, USA). The cleaned solutions were re-combined, dried down and re-suspended in 25 µL of 5 % FA prior to analysis by mass spectrometry.
iTRAQ-labeling of platelet digests
100 µg of digested platelet proteins were labeled with iTRAQ reagents (Applied Biosystems, Framingham, MA, USA). iTRAQ reagent 115 was resuspended in 70 µL of ethanol and added to the digested platelet sample. The pH was adjusted to between 7.5 and 8.5 using 0.5 M triethylammonium bicarbonate and the sample was incubated for 1 hour at room temperature. 100 µL of water was then added to quench the reaction and the iTRAQ labeled sample was dried down using a vacuum concentrator. The sample was re-suspended in Buffer A, acidified to below pH 3 and separated using the Ettan MDLC system as described in the previous paragraph.
Mass spectrometry
The MRM and MIDAS (MRM-initiated detection and sequencing) experiments were carried out on a 2000 Q-TRAP system (Applied Biosystems, Framingham, MA, USA) coupled to a nano-LC system (LC Packings, San Franscisco, CA, USA). 2 µl of platelet tryptic digests were separated over a 75 µm × 150 mm reversed-phase C18 column (particle size 3 µm, GL Sciences, Tokyo, Japan). A linear gradient of 5 to 60 % B in 50 min (solvent A: 95 % water + 0.1 % FA; solvent B: 80 % ACN + 0.1 % FA) at a flow rate of 200 nL/min was applied. Samples were analyzed on-line with nanospray ionization in positive mode. MRM was performed on predicted transitions determined using GPMDB’s MRM worksheet. MIDAS™ workflow designer software (Applied Biosystems) was used to calculate the collision energy for each individual peptide based on a linear relationship between precursor ion m/z and collision energy defined therein, and to generate the MIDAS acquisition methods. Typically, MRM transitions for at least three fragment ions per peptide were employed. Data were acquired for a minimum of three technical repeats and processed using Analyst 1.4.2. software (Applied Biosystems), and the correctness of the MRM-peptide assignment was confirmed by MIDAS experiments and database searches.
Results
Defining the information necessary for MRM assays
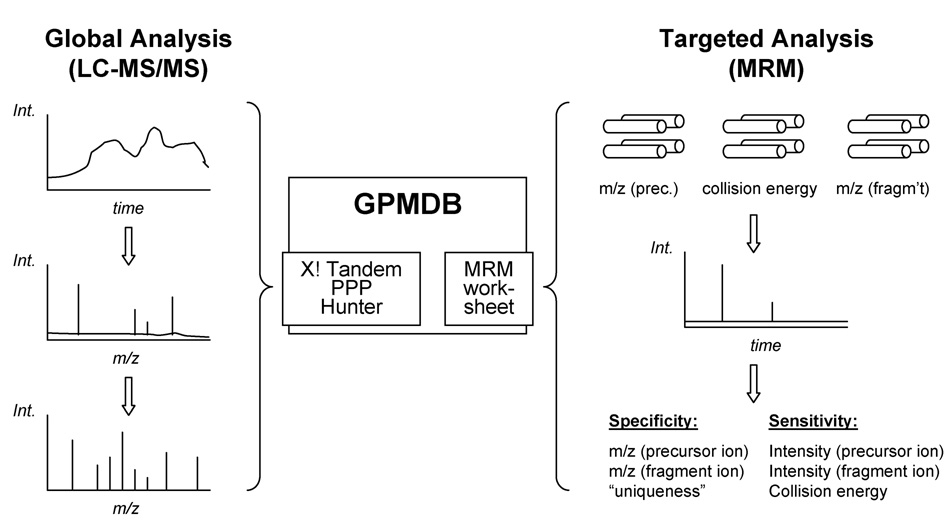
Multiple Reaction Monitoring (MRM) is commonly employed for the targeted analysis of drugs and metabolites due to its specificity and sensitivity. This is accomplished by operating the first and third quadrupole of a triple quadrupole-type instrument such that they let pass only ions with predetermined m/z while excluding all others (Fig. 1). While this results in the maximum specificity for the detection of a given analyte, the practical value of each transition will also depend on its “uniqueness” when applied to complex analyte mixtures. A transition that is shared by other analytes can lead to significant ambiguity in the subsequent peak assignment, and is often less valuable than a unique one, as additional, separation-specific information is then required.
Figure 1. Schematic outlining the data repository-driven approach for design of MRM transitions for targeted proteomics experiments.
The Global Proteome Machine Database (GPMDB) is the largest curated repository for peptide tandem mass spectra, which are generated by laboratories across the world, often using global proteomic approaches, and submitted for analysis using the X! Series search engines. A novel MRM worksheet was designed using the human data in the GPMDB and its utility as a tool for the design of MRM transitions was assessed experimentally. MRM experiments are conducted using a triple quadrupole-type instrument in which the first and third quadrupole are set up to only select ions with predetermined m/z ratios while excluding all others. Certain key pieces of information are required in order to successfully design specific and sensitive MRM transitions, including the m/z ratio and intensity of both the precursor and fragment ions, the collision energy and the “uniqueness” of the transition.
In contrast, the sensitivity of each transition is defined by the relative intensities of the precursor and fragment ion signals. While the former is a reflection of the physicochemical properties of the peptide and the ionization conditions, the latter depends on the stability of each bond in the peptide as well as the amount of energy imparted in the collision-induced dissociation (CID) step. Consequently, in addition to the four parameters contained in the survey and tandem mass spectra (m/z and intensity of precursor and fragment ions) available in each LC-MS/MS data set, two additional “soft” variables are needed to qualitatively and quantitatively describe an MRM assay: the collision energy that is associated with a given fragment ion intensity, and the uniqueness of the combination of precursor and fragment ion m/z values in a given sample. Only the m/z values and the collision energy are specified when the instrument is set up, however, with the collision energy effecting mostly the sensitivity of a transition, but not necessarily its specificity. Consequently, the m/z of precursor and fragment ion are the only parameters that are indispensable to set up and test initial MRM assays, while all other variables can be evaluated, validated and optimized in later stages, if necessary (Fig. 1).
Identifying MRM assay-relevant information in GPMDB
In the existing proteomics tools to design MRM transitions [9, 20], the four spectrum-related parameters and the two “soft” variables are predicted based on known information on process- or sample-related features (protein sequence and abundance, protease characteristics, physicochemical properties of amino acids, instrument type). To account for commonly observed deviations from theory, a higher than necessary number of transitions is initially predicted. These are subsequently tested to identify the most successful MRM assays, which are then further validated and optimized [26].
In order to test the hypothesis that extracting the relevant information from a spectrum libraries may represent an efficient shortcut for the design of MRM assays, the largest curated repository for tandem mass spectrometric data currently available was examined for the availability of the six previously mentioned variables. Three of the spectrum-based parameters were directly accessible in GPMDB: the m/z of the precursor ion, and the m/z and relative intensities of its characteristic fragment ions are stored for every entry in the archive. Conversely, the precursor ion intensity as the fourth spectrum-based parameter, as well as the two additional variables (collision energy and uniqueness), was not available.
While the available information would be sufficient to set up preliminary MRM assays for initial testing, this would only translate into a marginal improvement over the prediction approach due to the need of extensive evaluation. Alternative ways of defining these variables in GPMDB were thus investigated. For each precursor ion, the GPMDB only stores retention time, m/z ratio, peptide mass, charge state, and mass error as experimental information, none of which were suitable substitutes. Consequently, the recorded number of observations for each peptide across the entire repository was chosen as potential substitute for the precursor ion intensity, as this feature is also used for spectral counting in label-free quantitative proteomics [27]. Similarly, it was concluded that performing a comparative analysis of all database entries for their potential to produce the same combination of precursor and fragment ion m/z could be leveraged to determine uniqueness. In contrast, retrieving the optimal collision energy for a transition proved impossible, as this type of information is not retained in GPMDB. As the GPMDB defines consensus spectra for each peptide that are further sub-classified based on their differences in appearance, however, these can be utilized as “MRM patterns” that could potentially serve as valuable guides. This assumes that the operator and the majority of GPMDB contributors have optimized their individual instrument setups accordingly, as the relationship between collision energy and fragment ion intensity is highly dependent on the individual instrument settings [28].
Design of the MRM Worksheet in GPMDB
Based on these considerations, a design/testing/redesign cycle was employed to construct a user interface that could be utilized to produce testable MRM transitions based on the experimental results stored in GPMDB. This showed that there were four discrete sets of information that were required by the user.
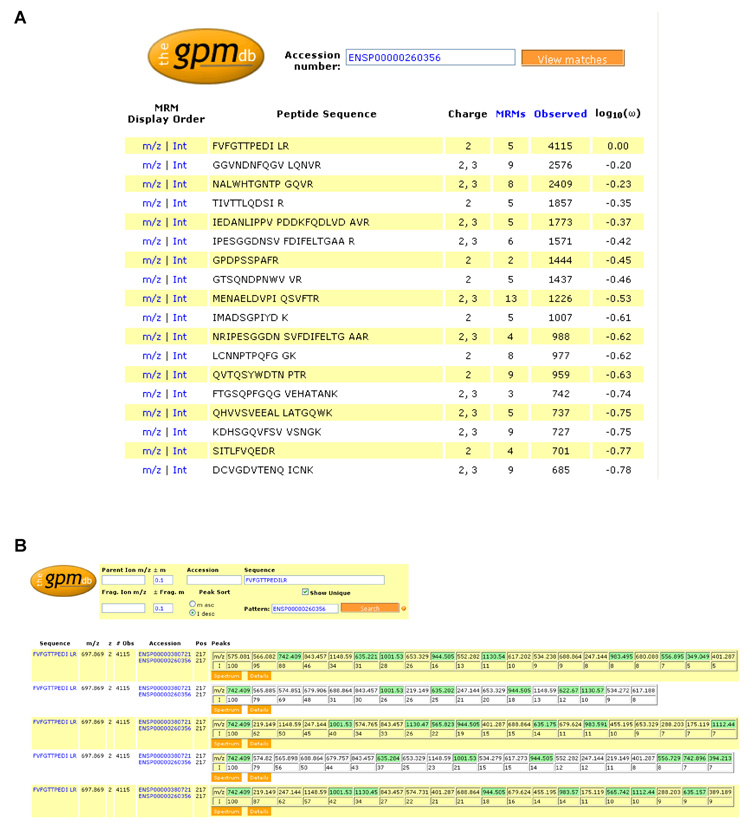
The first set of information was keyed to the sequence and accession number of the protein of interest. The information was rendered as a summary page of all of the peptides in a particular protein sequence that were candidates for an MRM assay. Information required on this page were the peptide sequence, known precursor ion charge states, the number of times each peptide had been observed, and the frequency of observation for each peptide (given in Fig. 2 as log10(ω), the decadic logarithm of the ratio of the number of observations of a particular peptide to the number of observations of the most frequently observed peptide for that protein, meanwhile changed to log10(ω/ω0) in GPMDB). Once this information was displayed, it was possible for an investigator to narrow their search to a few peptide sequences by inspection of the display (Fig. 2).
Figure 2. The GPMDB MRM worksheet.
Screen shots from the newly-developed worksheet for the design of MRM transitions at the http://gpmdb.thegpm.org. A. shows the MRM worksheet for the protein thrombospondin-1 (THBS1, swissprot accession number P07996), listing the peptide sequence, charge state the peptide has been observed in, the number of consensus spectra available for each peptide sequence across all charge states, sequence modifications, and instrument types (denoted as “MRMs” in the worksheet to appropriately reflect its use), the number of observations of the peptide in the database and the frequency of observation (log10(ω)). B. shows in detail the summary page displaying the 5 sets of MRMs predicted for the most observed thrombospondin-1 peptide (FVFGTTPEDILR), based on its 5 consensus spectra. The peptide sequence and occurring post-translational modification sites (indicated by the blue background and display of the associated mass shift when locating the mouse pointer on this position), m/z ratio, charge state and number of observations in the GPMDB of the precursor ion are listed, as well as the m/z ratio and relative intensities of the fragment ions. Unique transitions are highlighted in green. Note that the information in these screen shots was current as of 28th August 2008, but as the GPMDB is a dynamic, living database, certain information, such as the number of observations of a peptide, are likely to change with time as the database grows.
The next set of information was associated with the sequence of a particular peptide. A summary page containing all of the charge state, protein accession numbers and sequence modification information available for a particular peptide, as well as tables of fragment ion masses was sufficient for the user to judge which, if any, of the candidate spectra would contain transitions with the potential to be useful MRM transitions (Fig. 2).
Each of the potential candidate spectra could then be viewed as line spectrum histograms in a separate display already available in GPMDB, with the predictable b- and y- ions (as well as ammonia and water neutral loss ions) marked in color. Another display was designed to show the transitions from other peptides of the same precursor ion m/z that might cause interference signals that would impact the limits of detection and quantitation. This display also showed the interfering peptide sequences, charge states, HPLC relative retention times and the relevant protein accession numbers, Human Genome Naming Commission gene symbols and protein descriptions, when this information was available.
Testing the number of observations as substitute of peptide intensity
With this MRM worksheet in GPMDB in place, the impact of the substitutions and assumptions regarding the missing variables could be evaluated. To investigate whether the recorded number of peptide observations qualifies as a substitute for precursor ion intensity, several tryptic peptides originating from vinculin were picked such that they encompassed the entire scale of numbers of observations for this protein. Vinculin was chosen because it was consistently reported as the top-ranked protein in a series of previous global proteome analyses on similar platelet samples that had been carried out in the laboratory (data not shown). Subsequent MRM experimentation using the most intense fragment ion of each peptide, and determination of the peak area and signal-to-noise ratio for each of these transitions, provided a quantitative means of assessing the relationship between the number of observations and precursor ion intensity (Suppl. Fig. 1). While a noticeable increase in peak area and signal-to-noise ratio for higher numbers of observation was observed, a definitive correlation between either of the two parameters and the number of observations could not be established. Tentatively, a trendline was added to illustrate the increase, although it should be noted that there is currently no evidence for a linear relationship between these parameters.
Upon detailed inspection of the data points showing the largest deviation, two possible contributions to this outcome were identified. While some peptides were present in GPMDB only with one, others had been observed in two or three different charge states. Similarly, some peptides had been observed in their expected primary sequence only, while others had additional sequence modifications associated with them. In both cases, the number of observations contained in GPMDB reflected the net sum across all charge and modification states, making it impossible to discern the relative contribution of each species. For example, while the peptide AVAGNISDPGLQK was observed 2080 times, its entry in the MRM worksheet indicated ten different stored consensus patterns in GPMDB that were linked to 1+, 2+, and 3+ charge states and glutamine and an asparagine deamidation originating in the same peptide sequence, but not necessarily the same precursor ion m/z. Independent global proteome analysis of similar platelet samples had detected this peptide as doubly charged species in its non-modified and modified form.
Similarly, the peptide VMLVNSMNTVK was observed 991 times in the 2+ charge state, while the MRM worksheet indicated the existence of eight different stored consensus patterns in GPMDB, utilized as MRM patterns in the MRM worksheet in GPMDB, that corresponded to modified species after oxidation of one or two methionine residues, or asparagine deamidation. This peptide had also been detected as doubly charged, modified species with one or two methionine oxidations and deamidated asparagine in the independently performed global analyses, while a longer version of the peptide due to a missed cleavage of the carboxy-terminal lysine was also present. Conversely, the non-modified peptide form had not been detected at all in these untargeted studies.
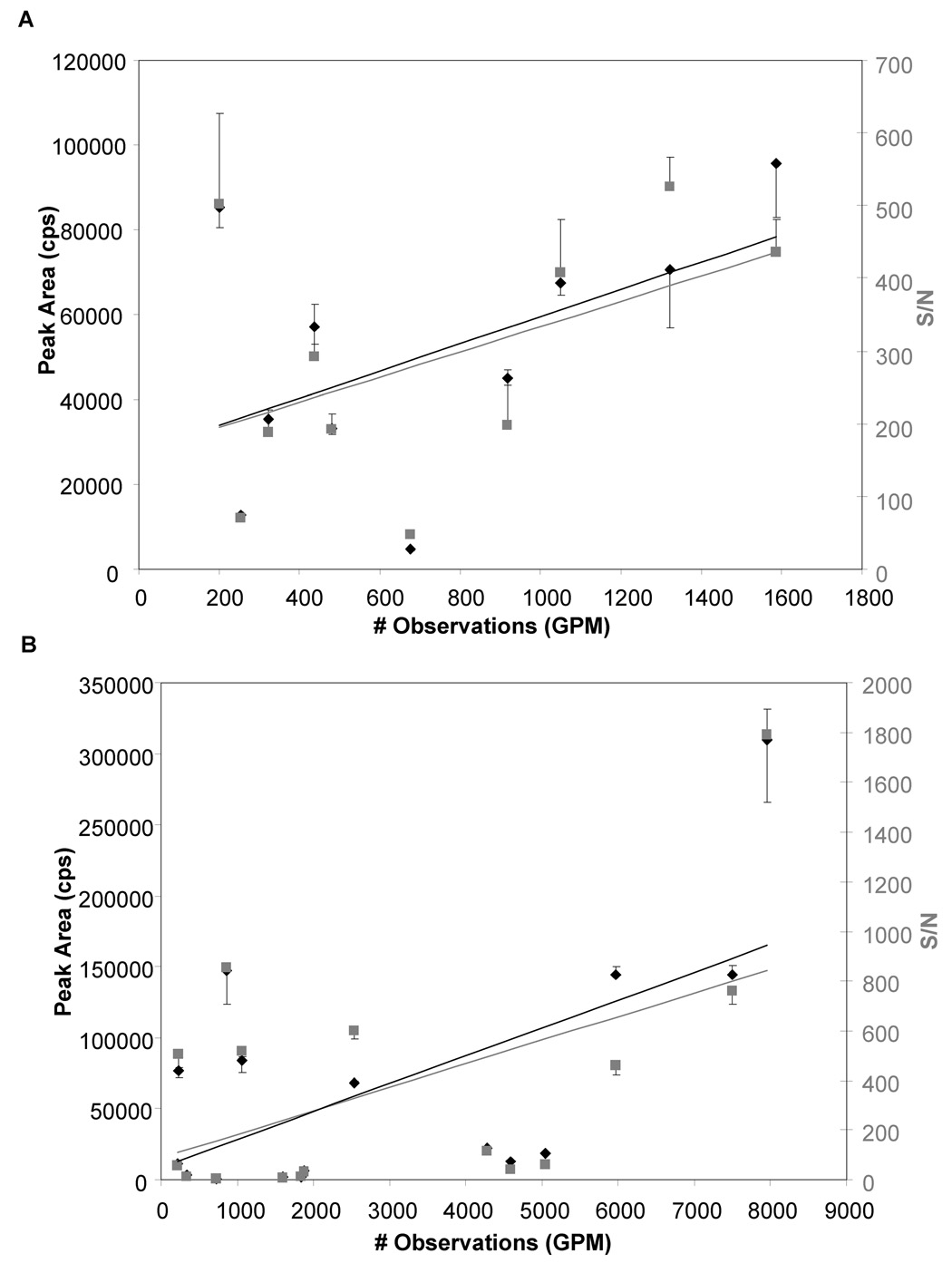
For a second protein, filamin A, a similar lack in correlation between the recorded number of observations and peak area or signal-to-noise was observed for a random selection of peptides (Suppl. Fig. 1). As the detection of multiple forms of a peptide in the same experiment results in increases in the number of observations while the individual peak area and signal-to-noise ratio decrease, this will generate data points that move towards the bottom right of the plot (Suppl. Fig. 1). In order to minimize such deviations, only vinculin- or filamin-derived tryptic peptides listed in the MRM worksheet as doubly charged species without any modifications were selected in a second series. Repeating the same data acquisition and processing steps then allowed reassessment of the relationship between the number of observations and precursor ion intensity (Fig. 3).
Figure 3. Peptide selection criteria affecting the usefulness of the number of observations in the GPMDB as an estimate of peptide abundance.
Scatter plots showing the number of peptide observations cited in the GPMDB vs the experimentally observed peak area (cps, left axis, black diamonds) and signal to noise ratio of the peaks (S/N, right axis, grey squares). Data shown is from GPM-predicted MRM transitions from 10 peptides from A. Vinculin (swissprot accession number P18206) and 16 peptides from B. Filamin A (swissprot accession number P21333), chosen as they had only been observed as doubly-charged species with no modifications in the GPMDB. While not all peptides followed the trend, in general an increased number of observations in the GPM correlated with higher peak areas and signal-to-noise ratios. Data is the mean of 3 replicate experiments + or − SD
Most notably, a much more significant correlation between the number of observations and MRM peak areas and signal-to-noise ratios was observed for both proteins when hypothesizing a linear relationship between the number of observations and peak area/signal-to-noise ratio, as apparent by the increase in r2-values obtained after linear regression from 0.08/0.10 (peak area/signal-to-noise ratio) to 0.26/0.23 for vinculin, and from 0.17/0.14 to 0.36/0.26 for filamin A. The second series of experiments supported the hypothesis that the number of observations is an adequate substitute for the precursor ion intensity, and underscored the limitations of the first series. There were still peptides that did not follow this trend, however, as apparent by the data points that deviate from the tentative trendline assuming a hypothetical linear relationship between these variables. They might be due to different yields of specific peptides across laboratories and variations in the ratio of abundances for select peptides originating in the same protein, which may be caused by differences in purity or activity of the digesting enzyme, the digestion conditions, or the peptide extraction or isolation protocols. Furthermore, variations in the experimental setup, such as exclusion of already sequenced peptides and the length of the exclusion time relative to the average chromatographic peak width, may have contributed to a non-linear relationship between the number of observations and peak area/signal-to-noise ratio. All of these possible sources of error cannot currently be evaluated, however, as the GPMDB does not archive the corresponding MIAPE (Minimum Information About a Proteomics Experiment) information.
Correlating collision energy with GPMDB consensus patterns
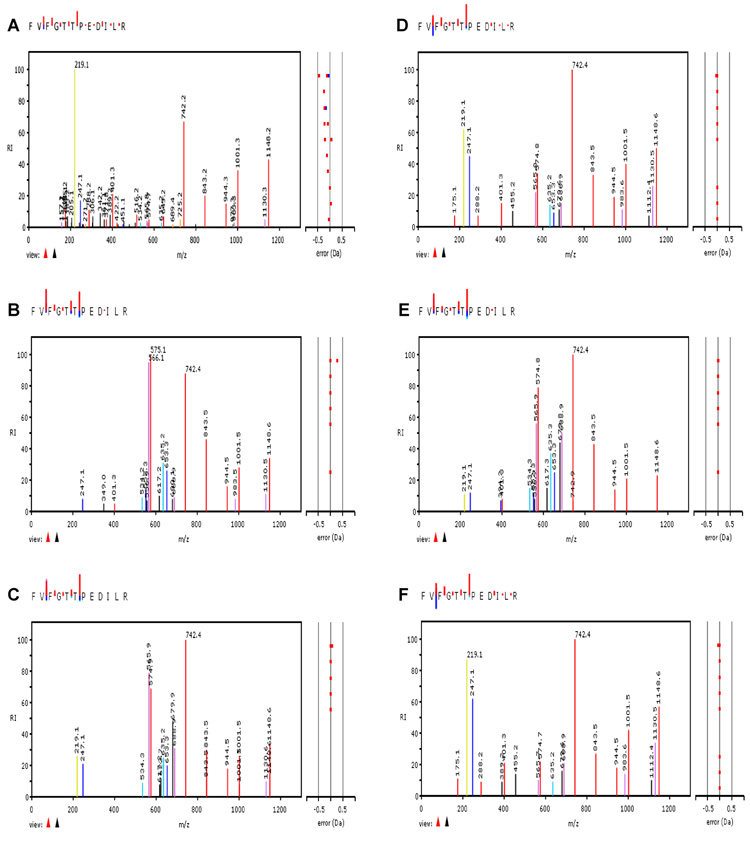
Next, the second variable contributing to the sensitivity of MRM transitions was investigated. To this end, the MIDAS (MRM-initiated detection and sequencing) workflow was employed, which performs MRM-triggered MS/MS experiments using collision energy settings that are calculated by the instrument software based on a linear dependence of the collision energy on the peptide m/z [9]. The resulting tandem mass spectra were then compared to the consensus patterns archived in GPMDB. As a representative example, the tandem mass spectrum of the most often observed peptide from the platelet protein thrombospondin, FVFGTTPEDILR, is shown in Fig. 4A, and the five consensus spectra in the GPMDB in Fig. 4B–F. While visual inspection already suggested good agreement between the experimental pattern and those present in GPMDB, cross-correlation analysis as a more quantitative readout confirmed this observation. For this peptide, correlation values between 0.755 and 0.893 were obtained, demonstrating the high similarity between these six spectra. Indeed, the experimental patterns appeared to be in good agreement with at least one of the consensus spectra for the majority of examples studied, as correlation values above 0.5 were easily obtainable, with values ranging between 0.8 and 0.9 in many cases. This supported the underlying assumption that for the majority of mass spectrometers employed in proteomic research, the collision energy is optimized, which ensures good agreement between experimental and consensus spectra without the need of storing and retrieving this information.
Figure 4. Correlation of collision energy and stored peptide consensus spectra.
MIDAS, which performs MRM-triggered MS/MS experiments based on previously optimized collision energy conditions, was conducted with the thrombospondin-1 peptide FVFGTTPEDILR to determine the correlation between collision energy and stored peptide fragmentation patterns. A. shows the fragment ion mass spectra experimentally acquired, which was compared with the average fragmentation patterns archived in the GPM (B–F).
For a more detailed comparison of experimental and archived consensus patterns, three technical repeats were carried out for three different peptides each of thrombospondin and actin (Table 1). For each peptide, five MRM transitions were set up that involved the two most intense unique and the three most intense non-unique fragment ions per peptide. The results indicated that the experimental fragment ion intensities were highly reproducible in all cases, excluding variation in the experimental setup as a possible source for the non-perfect correlation determined above. Moreover, the majority of fragment ions had intensities that differed by less than 50 % from those of the consensus spectra. In some cases, more significant deviations were present, e.g. for the y132+ fragment ion (m/z 706.4) of the actin peptide SYELPDGQVITIGNER and the a2 fragment ion (m/z 219.1) of the thrombospondin peptide FVFGTTPEDILR. This confirmed the general suitability of the GPMDB as a guide for the design of MRM transitions, but also highlighted its limitations in predicting the precise intensity distribution of fragment ions beyond a given tolerance.
Table 1. A comparison of GPM-archived fragment ion intensities and experimentally-determined fragment ion intensities in MRM transitions designed using the GPM database tool.
Table 1 lists GPM-predicted MRM transitions for several peptides from β-Actin (ACTB, swissprot accession number P60709) and Thrombospondin-1 (THBS1, swissprot accession number P07996), and compares fragment ion intensity values archived in the GPM vs experimentally determined average intensity values and SDs, based on 3 repeat experiments. Whether the peptide is considered unique by the GPM MRM prediction tool is noted in the final column
| Protein | Peptide Sequence | Q1 | Q3 | Fragment | Predicted | Average | SD | Unique? |
|---|---|---|---|---|---|---|---|---|
| m/z | m/z | Type | Intensity (GPM) | Intensity (Expt.) | ||||
| ACTB | AGFAGDDAPR | 488.7 | 630.3 | y6 | 100 | 100 | 0 | Y |
| 701.3 | y7 | 46 | 66.1 | 0.49 | Y | |||
| 343.2 | y3 | 61 | 23.8 | 0.53 | N | |||
| 424.7 | y8 | 33 | 34.6 | 0.57 | N | |||
| 276.1 | b3 | 16 | 60.1 | 1.72 | N | |||
| GYSFTTTAER | 566.8 | 437.2 | b4-H2O | 20 | 20.2 | 0.40 | Y | |
| 308.1 | b3 | 37 | 62.3 | 0.28 | Y | |||
| 678.3 | y6 | 100 | 100 | 0 | N | |||
| 912.4 | y8 | 77 | 53.6 | 2.10 | N | |||
| 577.3 | y5 | 50 | 52.9 | 0.22 | N | |||
| SYELPDGQVITIGNER | 895.9 | 1086.6 | y10 | 13 | 14.5 | 0.25 | Y | |
| 706.4 | y132+ | 30 | 83.1 | 0.72 | Y | |||
| 1298.7 | y12 | 100 | 46.1 | 0.16 | N | |||
| 475.2 | y4 | 65 | 100 | 0 | N | |||
| 689.4 | y6 | 62 | 95.1 | 1.71 | N | |||
| THBS1 | GFLLLASLR | 495.3 | 672.4 | y6 | 41 | 100 | 0 | Y |
| 318.2 | b3 | 18 | 85.0 | 5.47 | Y | |||
| 446.3 | y4 | 100 | 60.3 | 8.06 | N | |||
| 559.4 | y5 | 81 | 68.7 | 11.0 | N | |||
| 544.3 | b5 | 77 | 14.9 | 0.72 | N | |||
| FVFGTTPEDILR | 697.7 | 742.4 | y6 | 100 | 100 | 0 | Y | |
| 574.8 | y102+ | 79 | 84.9 | 0.40 | Y | |||
| 219.1 | a2 | 87 | 53.7 | 1.49 | N | |||
| 688.9 | y12-H2O2+ | 50 | 48.4 | 0.31 | N | |||
| 565.8 | y10-H2O2+ | 56 | 29.1 | 0.89 | N | |||
| GGVNDNFQGVLQNVR | 808.9 | 1083.5 | b11-H2O | 12 | 0 | 0.18 | Y | |
| 871.4 | b9-H2O | 12 | 1.6 | 0.51 | Y | |||
| 629.4 | y5 | 100 | 80.7 | 0.04 | N | |||
| 913.5 | y8 | 47 | 50.8 | 0.48 | N | |||
| 785.5 | y7 | 62 | 100 | 0 | N |
Assessing the uniqueness of MRM transitions
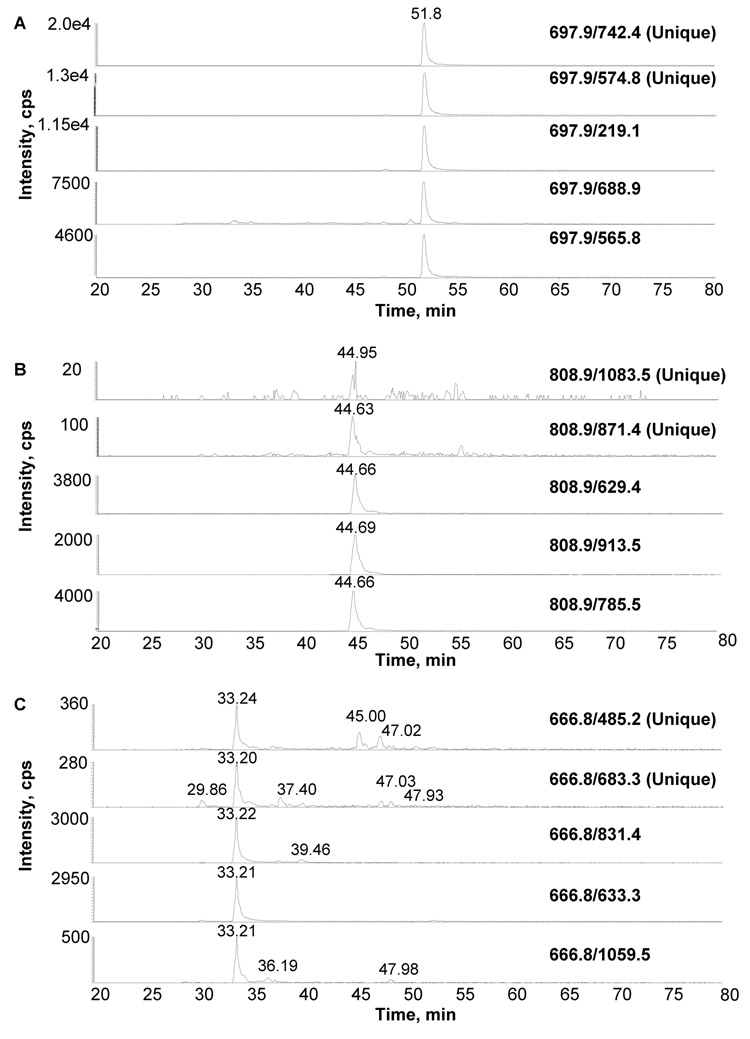
In order to determine whether the truthful prediction of unique transitions based on the data stored in GPMDB is possible, the MRM worksheet was set up so it could highlight transitions predicted to be unique within a given mass window for each peptide. For a significant number of peptides, two such predicted unique transitions were monitored together with three non-unique ones, and their MRM traces were analyzed for the peak areas of the designed transition as well as any additional ones that would be present (Suppl. Table 1). While in several cases unique transitions indeed resulted in a single peak in the MRM trace, those were restricted to transitions involving high intensity fragment ions, as seen in the example of the thrombospondin peptide FVFGTTPEDILR (Fig. 5A). Conversely, predicted unique transitions based on fragment ions associated with low intensity, or originating in secondary fragmentation, were typically linked to lower signal-to-noise ratios as well as additional signals in the MRM trace. Examples for this observation are the thrombospondin peptides GGVNDNFQGVLQNVR (Fig. 5B) and LCNNPTPQFGGK (Fig. 5C), for which the two most intense unique fragment ions were predicted to be b11-H20 and b9-H20, and b4-NH3 and b6-NH3, respectively (Suppl. Table 1). Overall, the relative intensity of the fragment ion fared significantly better as an indicator for the quality of a transition than the predicted uniqueness in these studies, as exemplified by the remaining MRM traces of these two peptides.
Figure 5. Assessment of the importance of the uniqueness of the transition when designing MRM experiments.
Two unique transitions together with three non-unique transitions were examined for a number of peptides from β-actin and thrombospondin-1 (see table 2 for a complete list). Shown are representative MRM tracings from three scenarios that were observed. A. shows MRM traces from peptide FVFGTTPEDILR, (thrombospondin-1) showing high intensity, specific signals for both unique and nonunique transitions. In contrast, B. shows MRM traces from the peptide GGVNDNFQGVLQNVR (thrombospondin-1) showing that the 2 transitions predicted to be unique did not prove to be useful as they were of much lower intensity then the nonunique peptides. Finally, C. shows MRM traces from the peptide LCNNPTPQFGGK, (thrombospondin-1) showing again that the unique transitions proved to be misleading as they resulted in low intensity signals and the presence of secondary fragment ions, while the transitions predicted to be non-unique proved to have higher intensity signals with less interference.
Predicting MRM transitions of isotope-labeled peptides
A final series of experiments involving various quantitative proteomics approaches was performed to test the usefulness of the new MRM worksheet for the design of MRM assays that involve the relative or absolute quantitation of the analyte. Standard dilution series experiments indicated that label-free quantitation based on the ratio of MRM peak areas of the analyte relative to internal standards was sufficient to estimate the relative protein abundance across samples (data not shown). Additional independent experiments suggested that metabolic or chemical labeling via incorporation of stable isotope coded amino acids into proteins (SILAC) or synthetic peptides (AQUA) would not lead to noticeable fragmentation pattern changes.
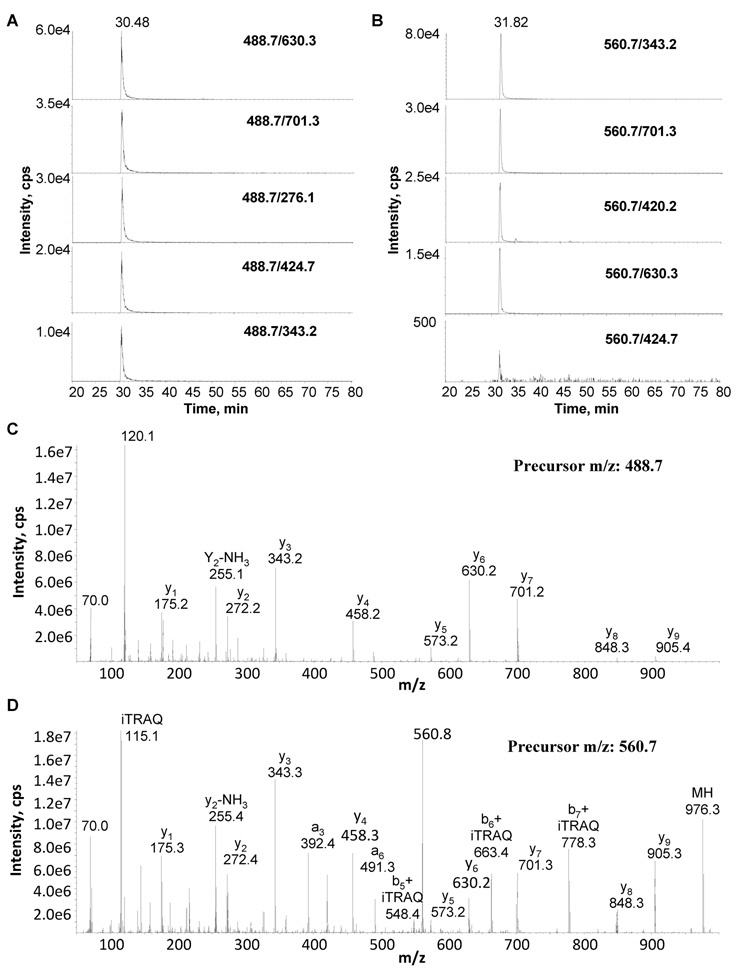
Conversely, chemical labeling via the addition of different isotope-coded functional groups to peptides could have a profound effect on the appearance of tandem mass spectra. While the chemistry of amine-reactive compounds such as the iTRAQ or mTRAQ reagents allows for an easy adjustment of the m/z values of the precursor and fragment ions based on the known number of reactive sites each peptide contains, their impact on the fragmentation patterns and the fragment ion intensities is less predictable as they alter the distribution of basic moieties in the peptides [29]. To determine whether the MRM worksheet in GPMDB can also be utilized for the design of MRM transitions of iTRAQ-labeled peptides, tryptic digests of platelet lysates were reacted with the iTRAQ reagent 115. The results of the MRM assays carried out before labeling were then compared to those obtained after iTRAQ labeling and correction of the m/z and collision energy values used in the MRM transitions, calculated based on the mass shifts imparted by the incorporation of the iTRAQ reagent and the resulting increase in peptide mass and precursor ion m/z, respectively. Changes in the relative peak areas of the different MRM transitions were commonly seen despite the adjustment of the collision energy upon iTRAQ labeling, as evident for the actin peptide AGFAGDDAPR (Fig. 6). No significant loss in specificity due to iTRAQ labeling and transition adjustment was noticed, however. This implies that the changes resulted in altered fragment ion intensities without significantly affecting the overall fragment ion formation, albeit some doubly-charged fragment ions disappeared (Suppl. Table 2). Indeed, the changes in sensitivity correlated well with the altered fragment ion intensities seen in the individual tandem mass spectra upon labeling, as the iTRAQ label commonly enhanced the signal intensities of b-ion series without significantly affecting the y-ion series (Fig. 6). While this demonstrated the general usefulness of the MRM worksheet in GPMDB also for quantitative proteomics applications involving iTRAQ chemistry, it highlighted the need to adjust the GPMDB-based MRM transitions already in the initial evaluation step, rather than during subsequent validation and optimization.
Figure 6. Design of MRM transitions using iTRAQ-labeled samples.
Figure 6A shows MRM traces for the ion pairs as selected from the GPM. All five ion pairs chosen show enough intensity for positive identification of the desired peptide, in this case AGFAGDDAPR from β-actin. Figure 6B shows the MRM traces for the same ion pairs for the same β-actin peptide labeled with iTRAQ. The relative position of the intense ions pairs has changed compared to the non-labeled ion pairs, but overall intensities remain strong to give positive identification of the desired labeled peptide. Figure 6C and D are a comparison of MS/MS spectra of non-labeled and iTRAQ-labeled AGFAGDDAPR from β-actin. Figure 6C shows the MS/MS spectrum of non iTRAQ-labelled peptide AGFAGDDAPR from β-actin and figure 6D shows the MS/MS spectrum of the same peptide labelled with iTRAQ. The y-fragment ions from 1 to 9 are present in both the non-labelled and labelled spectra with different relative intensities to each other. The b-fragment ions gain more intensity and their presence is easier to detect with the iTRAQ labelled peptide.
Discussion
The targeted analysis of individual proteins, often after an initial discovery phase, is a key step in proteomics. MRM assays are particularly useful as they provide the high sensitivity, specificity, and throughput that is necessary to carry out large numbers of experiments. The new MRM worksheet within the GPMDB framework represents a universal interface for the design of targeted MRM studies. As it is based on communal evidence rather than theoretical predictions or previously performed own experiments, it can provide an efficient shortcut to the design of MRM assays for tryptic peptides of human proteins, as demonstrated here.
Initial assessment showed that three of the six variables needed to define an MRM assay are readily accessible in GPMDB. Conversely, the precursor ion intensity and the collision energy associated with a given fragment ion intensity are not available. In fact, retaining this type of information in GPMDB would significantly impair its performance. Recording the absolute precursor ion intensity would require archiving all survey mass spectra of each experiment, thus creating dramatically larger data sets. At the same time, the comparative value of such information would be limited. Indeed, the well-known limited reproducibility of LC-MS experiments even for technical replicates on the same platform has spurred the development of computational solutions to compensate for them, including for MRM assays [30]. Across platforms, laboratories, and samples, the differences would be heightened further and far outweigh the potential similarities of patterns. Consequently, the GPMDB intentionally only archives the minimum amount of LC-MS information linked with each tandem mass spectrum: the relative position in the chromatogram (spectrum number), the precursor ion m/z, and the deduced charge state, peptide mass, and mass error upon peptide sequence assignment.
Similarly, the collision energy used to generate a tandem mass spectrum can vary dramatically across platforms and laboratories. In addition, other instrumental and experimental variables such as collision gas density and purity or the total number of ions contained in the collision cell can have a significant influence on the resulting fragmentation patterns. As keeping track of all of these variables would be an insurmountable task, the GPMDB has taken a reductionistic approach by archiving only the most significant features of each tandem mass spectrum: a maximum of 50 most intense signals present with at least 1 % of the base peak intensity. These are then grouped into consensus spectra based on their similarities. While this implies that ultimately the onus is on the experimenter to ensure optimal fragmentation in the respective individual analyses submitted to the GPMDB, the impact of individual suboptimal data is minimized by an averaging effect when a large number of laboratories contribute. Indeed, the more often a particular peptide is observed, the more likely a statistical assignment to a consensus pattern or the recognition of a different, not previously observed pattern becomes. This is directly reflected in the number of consensus/MRM patterns stored for each peptide sequence in GPMDB.
The absence of records on the precursor ion intensity and the collision energy associated with a given fragment ion intensity in GPMDB translates into the potential loss of sensitivity for individual MRM assays. As the validation and optimization of experimental conditions is a common step in MRM applications to maximize their sensitivity, having adequate substitutes to estimate precursor ion intensity and collision energy will be sufficient to test and evaluate the initial setup. As demonstrated here, the number of observations and the recorded consensus spectra are such substitutes. Replacing the precursor ion intensity in individual experiments with the total number of observations across all experiments enables the prediction of MRM transitions likely to result in high peak areas and signal-to-noise ratios. In this study, no significant difference was observed between peak area and signal-to-noise ratio. Exceptions from the general trend do still exist, which needs to be taken into account when setting up the initial MRM screens.
Similarly, careful optimization of the collision energy on the instrument chosen for the MRM assays is sufficient to facilitate the use of the consensus patterns stored in GPMDB to predict fragment ion intensities. This also suggests that these archived patterns are indeed compiled from individual spectra that were obtained at optimized collision energy settings. However, caution is necessary when working with peptides that have been observed as a number of species, be they due to the presence of post-translational modifications, different charge states, or different consensus spectra. As the number of observations is cumulative across all species and experimental patterns, individual intensities can deviate significantly from the general trend in such cases. Strikingly, the results presented here provide experimental proof for some of the “common sense” rules that have been reported for the design of MRM transitions [9, 26]. These include, for example, avoiding peptides that contain potential modification sites, are prone to missed cleavages, or produce several charge states, unless one decides to monitor all of these species instead. The MRM worksheet lists the charge state(s) and number of MRM patterns underlying the total number of observations. These can be utilized as a guide to identify such cases based on experimental evidence rather than theoretical considerations. It is important to bear in mind, however, that successfully setting up the initial MRM experiments does not eliminate the need for subsequent validation and optimization, as significant deviations from the information contained in the GPMDB indeed occur.
The sixth variable that is relevant for the design of MRM assays, the “uniqueness” of a transition, is associated with the identification of additional precursor ion/fragment ion combinations that could interfere with the detection of a given peptide. It is key to the specificity of the detection, the confidence in the assignment, and the throughput that can be achieved in a single experiment. Indeed, monitoring a single unique transition per peptide would allow increasing the total number of peptides studied in a single LC-MS experiment without changing the individual or overall dwell time or sacrificing confidence. The GPM database is a tremendous resource for the identification of real rather than predicted interferences, as it holds a vast array of experimental observations. For each peptide, it highlights the fragment ions that result in an MRM transition that has not been observed for any other peptide. Still, relying on predicted unique transitions had only limited success for the test samples.
Two possible factors may have led to this outcome. Any information that is not stored in GPMDB is not considered for the prediction of uniqueness. Peptides that have not been sequenced before, e.g. due to low intensity or the presence of uncommon modifications, and all peptides that result in non-assignable fragment mass spectra are absent from GPMDB. Moreover, the rate of failed assignment is highly dependent on the instrument type, and varies between 20 % and 85 % of the submitted tandem mass spectra. Similarly, the m/z of low intensity fragment ions may not be retained, as the GPMDB only stores the 50 most intense signals in each tandem mass spectrum, provided they show at least 1 % of the base peak intensity. As a result, the information underlying the prediction of uniqueness could indeed be biased towards standard experimental features. In contrast, individual experiments that use conditions deviating from those typically used in the field may result in a different proteome composition that would limit the success of this prediction. Choosing samples derived from platelets, which are anucleate cell fragments, may have contributed to the limited success of the “uniqueness” feature in this study.
Secondly, the sensitivity of the detection may also play a role. The peak area obtained in MRM assays is a function of the precursor and fragment ion intensities as well as the applied collision energy. In the course of this study, it was noticed that unique transitions are often associated with less common fragment ions, e.g. multiply charged fragment ions, secondary or internal fragment ions (see Suppl. Table 1 and Fig. 5), or even signals that could not easily be assigned to the peptide sequence. In the majority of cases, these are present in the tandem mass spectrum with relative intensities below 20% of the base peak, reducing the peak area and sensitivity compared to MRM transitions relying on intense fragment ions in non-unique combinations. For abundant peptides, such a loss in sensitivity is often tolerable. For less abundant peptides, however, the lower precursor ion intensity reduces the MRM sensitivity and increases the potential of failed detection in complex mixtures. In these cases, it may be more appropriate to rely on several non-unique but intense transitions to overcome limited sensitivity of unique transitions, at the expense of reducing the number of peptides that can be monitored per experiment. As this currently is common practice for MRM experimentation, the “uniqueness” prediction provides an additional feature that is not otherwise available. The MRM worksheet is designed to provide the maximum amount of information to support the user in making decisions. A link “Details” has been added to display specific information of potentially interfering peptides, including relative fragment ion intensity and estimated retention time difference. While these additional features can provide valuable support, it is ultimately at each user’s discretion to define their own priorities and when testing and optimizing their experimental setup. In this respect, utilizing the LC retention time is particularly promising. As this parameter is highly dependent upon, and will vary significantly with, the HPLC system and setup that is used, its detailed investigation was beyond the scope of this study, however.
Finally, the ease and robustness of the prediction of MRM transitions of iTRAQ-labeled peptides underscores the practical value of this MRM prediction tool. As only a limited number of reference spectra exist for this type of sample, GPMDB cannot currently support the direct prediction of suitable transitions. Straightforward adjustment of the m/z values of the precursor and fragment ion and the collision energy was shown to be sufficient to correct for the experimental change to a large extent, albeit the absolute fragment ion intensities did no longer match those recorded in the GPMDB. Optimization of the most successful MRM transitions, generally carried out once preliminary MRM assays have been tested, would easily compensate for such changes, if necessary. The MRM worksheet of GPMDB therefore also represents a powerful tool for the initial selection of candidate MRM transitions in quantitative proteomics.
It should be noted that this proof-of-concept study was performed on tryptic peptides derived from human proteins. They represent the largest contingent of tandem mass spectra in the GPMDB, and are readily accessible by searching the GPMDB for the protein name or its accession number in the user interface. As the data volumes involving other organisms rapidly expand, the design of MRM assays for additional organisms based on GPMDB may well be feasible. Researchers may query GPMDB or contact the authors for more detailed information.
Supplementary Material
Scatter plot showing the number of peptide observations cited in the GPMDB vs the experimentally observed peak area (cps, left axis, black diamonds) and signal-to-noise ratio of the peaks (S/N, right axis, grey squares). Data shown is from GPM-predicted MRM transitions from 8 peptides from A. vinculin (swissprot accession number P18206) and 8 peptides from B. filamin A (swissprot accession number P21333). While there is a general increase in peak area and signal-to-noise ratio with increasing number of observations in the GPMDB, a strong correlation was not observed. Data is the mean of 3 replicate experiments + or − SD
Acknowledgements
This work was funded by operating grants from the Canadian Blood Services (JK) and the National Institutes of Health (RCB). GMW is supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research/Heart and Stroke Foundation of Canada (CIHR/HSFC) Strategic Training Program in Transfusion Science through the Centre for Blood Research (CBR) at the University of British Columbia. This work was carried out in the Multi-user Facility for Functional Proteomics at the Biomedical Research Centre, which is supported by infrastructure funding from the Michael Smith Foundation for Health Research (MSFHR). The authors would like to thank Jenn Yuen for phlebotomy services, conducted at the CBR Research Blood Collection Suite, and Jason Rogalski for technical advice and general support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;vol. 422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;vol. 312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 3.Malmstrom J, Lee H, Aebersold R. Advances in proteomic workflows for systems biology. Curr Opin Biotechnol. 2007;vol. 18:378–384. doi: 10.1016/j.copbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang N, Li L. Exploring the precursor ion exclusion feature of liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry for improving protein identification in shotgun proteome analysis. Anal Chem. 2008;vol. 80:4696–4710. doi: 10.1021/ac800260w. [DOI] [PubMed] [Google Scholar]
- 5.Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics. 2007;vol. 6:1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;vol. 8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Higton DM. A rapid, automated approach to optimisation of multiple reaction monitoring conditions for quantitative bioanalytical mass spectrometry. Rapid Commun Mass Spectrom. 2001;vol. 15:1922–1930. doi: 10.1002/rcm.454. [DOI] [PubMed] [Google Scholar]
- 8.Gergov M, Ojanpera I, Vuori E. Simultaneous screening for 238 drugs in blood by liquid chromatography-ion spray tandem mass spectrometry with multiple-reaction monitoring. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;vol. 795:41–53. doi: 10.1016/s1570-0232(03)00498-7. [DOI] [PubMed] [Google Scholar]
- 9.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;vol. 5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;vol. 6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed N, Thornalley PJ. Quantitative screening of protein biomarkers of early glycation, advanced glycation, oxidation and nitrosation in cellular and extracellular proteins by tandem mass spectrometry multiple reaction monitoring. Biochem Soc Trans. 2003;vol.31:1417–1422. doi: 10.1042/bst0311417. [DOI] [PubMed] [Google Scholar]
- 12.Cox DM, Zhong F, Du M, Duchoslav E, Sakuma T, McDermott JC. Multiple reaction monitoring as a method for identifying protein posttranslational modifications. J Biomol Tech. 2005;vol. 16:83–90. [PMC free article] [PubMed] [Google Scholar]
- 13.Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics. 2005;vol. 4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Ciccimaro E, Hevko J, Blair IA. Analysis of phosphorylation sites on focal adhesion kinase using nanospray liquid chromatography/multiple reaction monitoring mass spectrometry. Rapid Commun Mass Spectrom. 2006;vol. 20:3681–3692. doi: 10.1002/rcm.2783. [DOI] [PubMed] [Google Scholar]
- 15.Hulsmeier AJ, Paesold-Burda P, Hennet T. N-glycosylation site occupancy in serum glycoproteins using multiple reaction monitoring liquid chromatography-mass spectrometry. Mol Cell Proteomics. 2007;vol. 6:2132–2138. doi: 10.1074/mcp.M700361-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Janecki DJ, Bemis KG, Tegeler TJ, Sanghani PC, Zhai L, Hurley TD, Bosron WF, Wang M. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal Biochem. 2007;vol. 369:18–26. doi: 10.1016/j.ab.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 17.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;vol. 104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSouza LV, Taylor AM, Li W, Minkoff MS, Romaschin AD, Colgan TJ, Siu KW. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J Proteome Res. 2008;vol. 7:3525–3534. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- 19.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;vol. 25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 20.Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics. 2008;vol. 7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;vol. 20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 22.Craig R, Cortens JP, Beavis RC. The use of proteotypic peptide libraries for protein identification. Rapid Commun Mass Spectrom. 2005;vol. 19:1844–1850. doi: 10.1002/rcm.1992. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res. 2006;vol. 5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 24.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;vol. 3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 25.Krokhin OV, Craig R, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA. An improved model for prediction of retention times of tryptic peptides in ion pair reversed-phase HPLC: its application to protein peptide mapping by off-line HPLC-MALDI MS. Mol Cell Proteomics. 2004;vol. 3:908–919. doi: 10.1074/mcp.M400031-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Han B, Higgs RE. Proteomics: from hypothesis to quantitative assay on a single platform. Guidelines for developing MRM assays using ion trap mass spectrometers. Brief Funct Genomic Proteomic. 2008 doi: 10.1093/bfgp/eln032. [DOI] [PubMed] [Google Scholar]
- 27.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;vol. 4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Rogalski JC, Lin MS, Sniatynski J, Taylor RJ, Youhnovski N, Przybylski M, Kast J. Statistical evaluation of electrospray tandem mass spectra for optimized peptide fragmentation. J Am Soc Mass Spectrom. 2005;vol. 16:505–514. doi: 10.1016/j.jasms.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Locke SJ, Leslie AD, Melanson JE, Pinto DM. Deviation from the mobile proton model in amino-modified peptides: implications for multiple reaction monitoring analysis of peptides. Rapid Commun Mass Spectrom. 2006;vol. 20:1525–1530. doi: 10.1002/rcm.2512. [DOI] [PubMed] [Google Scholar]
- 30.Martin DB, Holzman T, May D, Peterson A, Eastham A, Eng J, McIntosh M. MRMer: An interactive open-source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M700504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plot showing the number of peptide observations cited in the GPMDB vs the experimentally observed peak area (cps, left axis, black diamonds) and signal-to-noise ratio of the peaks (S/N, right axis, grey squares). Data shown is from GPM-predicted MRM transitions from 8 peptides from A. vinculin (swissprot accession number P18206) and 8 peptides from B. filamin A (swissprot accession number P21333). While there is a general increase in peak area and signal-to-noise ratio with increasing number of observations in the GPMDB, a strong correlation was not observed. Data is the mean of 3 replicate experiments + or − SD