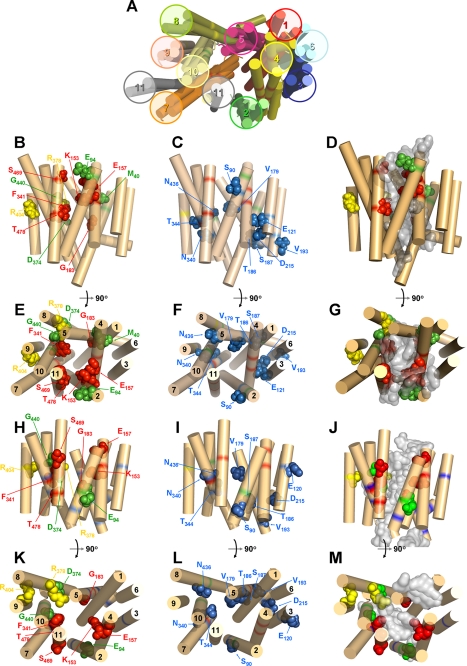
FIGURE 1.
Structural models of LdNT1.1 based on ab initio and threading analyses. Helices are indicated by rigid cylinders and are numbered 1–11. Specific amino acid side chains are shown as space-filling models and are labeled with the corresponding residue designation. A, three hypothetical ab initio models for LdNT1.1 derived from Rosetta modeling are presented and compared with a model obtained by threading analysis upon the template of the 3-glycerol-phosphate transporter of E. coli. B, C, E, and F depict side and cytosolic views, respectively, of preferred ab initio model 2. H, I, K, and L depict side and cytosolic views, respectively, of the threading model. Residues whose mutation cripples transport are highlighted in green, residues whose mutation alters substrate specificity are colored in red, residues whose mutation disrupts trafficking to the cell surface are shown in yellow, and residues whose mutation produces moderate changes in transport activity are separately grouped (C, F, I, and L) and depicted in blue. Mutations in several residues highlighted here, Glu121 and Asp215 (9) and Thr186 and Val193 (13), were not reported elsewhere in this manuscript but were documented in the associated references. D, G, J, and M offer side and cytosolic views, respectively, of the docking of adenosine (gray density) into the ab initio (D and G) and threading models (J and M). The structures in this figure and in Fig. 4 were generated using PyMOL.