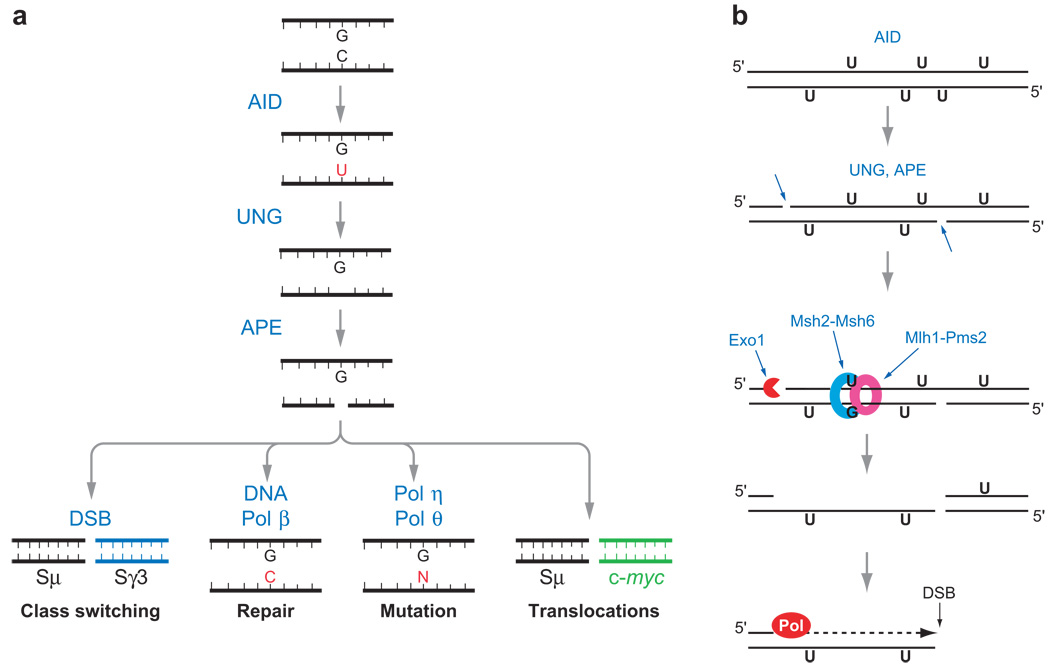
Figure 2.
Diagrams of (a) model for generation of DNA breaks, mutations, and translocations in IgS regions by AID-UNG-APE and (b) model for conversion of SSBs to DSBs by mismatch repair. (a) AID deaminates dC, resulting in dU bases, which are excised by one of the uracil DNA glycosylases, UNG. Abasic sites are cut by AP-endonuclease (APE1 and APE2) (74), creating SSBs that can spontaneously form DSBs if they are near each other on opposite DNA strands, or, if not, mismatch repair activity converts them to DSBs (see b). Alternatively, DNA Pol β can correctly repair the nick, preventing CSR, or error-prone translesion polymerases can repair the nick but introduce mutations. Finally, the DNA breaks can lead to aberrant recombinations/translocations. (b) AID is hypothesized to introduce several dU residues in S regions during one cell cycle. Some of the dU residues are excised by UNG, and some of the abasic sites are nicked by APE. The U:G mismatches that remain would be substrates for Msh2-–Msh6 (100). Msh2-–Msh6, along with Mlh1-Pms2, recruit Exo1 (and accessory proteins) to a nearby 5′ nick, from where Exo1 begins to excise toward the mismatch (90, 91), creating a DSB with a 5′ single-strand overhang, which can be filled in by DNA polymerase. Fill-in synthesis is probably performed by translesion polymerases owing to the presence of abasic sites. Alternatively, the overhang is removed by a 5′ flap endonuclease (Fen1) or by Exo1.