Abstract
Background and Aims
Callose involvement in spore development is a plesiomorphic feature of land plants. Correlated light, fluorescence and immuno-electron microscopy was conducted on the developing spores of Physcomitrella patens to probe for callose. Using a bioinformatic approach, the callose synthase (PpCalS) genes were annotated and PpCalS and AtCalS gene families compared, testing the hypothesis that an exine development orthologue is present in P. patens based on deduced polypeptide similarity with AtCalS5, a known exine development gene.
Methods
Spores were stained with aniline blue fluorescent dye. Capsules were prepared for immuno-light and immuno-electron microscopy by gold labelling callose epitopes with monoclonal antibody. BLAST searches were conducted using the AtCalS5 sequence as a query against the P. patens genome. Phylogenomic analysis of the CalS gene family was conducted using PAUP (v.4·1b10).
Key Results
Callose is briefly present in the aperture of developing P. patens spores. The PpCalS gene family consists of 12 copies that fall into three distinct clades with AtCalS genes. PpCalS5 is an orthologue to AtCalS5 with highly conserved domains and 64 % similarity of their deduced polypeptides.
Conclusions
This is the first study to identify the presence of callose in moss spores. AtCalS5 was previously shown to be involved in pollen exine development, thus making PpCalS5 a suspect gene involved in moss spore exine development.
Key words: Bryophyte, callose, callose synthase, exine development, moss, Physcomitrella patens, spores, sporogenesis
INTRODUCTION
Spores are among the primary adaptations that enabled ancient land plants to survive in environmentally harsh landscapes (Graham, 1993; Shaw and Renzaglia, 2004; Blackmore, 2007; Renzaglia et al., 2007). Early land plant radiation was coupled with elaboration from a single-celled zygote in the Charales to the multicellular sporophyte generation, which amplified spore production, but preceded seed evolution (Renzaglia et al., 2007). Unicellular meiospores function as key perennating and dispersal units by producing complex walls that are resistant to microbial damage and decay. Within embryophytes, multilayered spore walls are systematically laid down in a prescribed manner. While it is generally accepted that bryophyte spores have an intine of cellulose that is surrounded by an exine impregnated with sporopollenin, precise identification of spore wall constituents is lacking.
As part of the Green Tree of Life Project to describe and compare the process of sporogenesis across bryophytes (mosses, liverworts, hornworts), ontogenetic changes in spore wall constituents were examined in the model moss Physcomitrella patens. A novel finding in this study was the discovery of callose in the aperture exine. While involvement of callose in sporogenesis of the green alga Coleochaete and liverworts is well-documented, to date this polysaccharide has not been detected in association with sporogenesis in any moss (Neidhart, 1979; Graham and Taylor, 1986; Brown and Lemmon, 1990).
Callose is a linear 1,3-β-glucan molecule that is widely distributed in the embryophyte cell wall. This transient polysaccharide is involved in fundamental biological processes and is considered to be evolutionarily important in land plants (Aspinall and Kessler, 1957; Graham et al., 1991; Stone and Clarke, 1992; Garbary and Renzaglia, 1998; Hong et al., 2001; Scherp et al., 2001; Verma and Hong, 2001; Ligrone et al., 2002). In angiosperms callose is necessary for proper pollen wall formation and patterning (Dong et al., 2005; Nishikawa et al., 2005), cell plate formation (Stone and Clarke, 1992; Hong et al., 2001; Geisler-Lee et al., 2002; Jacobs et al., 2003), sieve plate production in phloem, megasporogenesis, microsporogenesis and wound healing (Scherp et al., 2001; Verma and Hong, 2001; Evert and Eichhorn, 2006). Callose is reported to guide spore wall development in heterosporous lycopsids (Lugardon, 1990; Gabarayeva and Hemsley, 2006). In bryophytes, callose surrounds young spermatids during development and is associated with modification of plasmodesmata in nacreous leptoid pores (Gorska-Brylass, 1969; Stevenson, 1974; Graham et al., 1991; Graham, 1993; Garbary and Renzaglia, 1998). Additionally, curdlans (callose-like molecules) are present in bacteria, and the genes responsible for the sucrose synthesis pathway necessary for callose synthesis are in cyanobacteria, suggesting this process is deeply conserved and is likely to have evolved specialized functions within the various groups of organisms (Scherp et al., 2001; Verma and Hong, 2001; McIntosh et al., 2005; Curatti et al., 2006).
The availability of whole genome sequences affords the opportunity to explore the evolution of function and structure of genes across land plants. A number of P. patens gene families have been annotated and characterized including peptidoglycan biosynthesis (Machida et al., 2006), inorganic phosphate (Pi) uptake (Wang et al., 2008), aldehyde dehydrogenases (Wood and Duff, 2009) and cellulose synthase (Roberts and Bushoven, 2007). The production of callose via the callose synthase (CalS a.k.a. GSL 1,3-beta-glucan synthase EC 2·4·1·34) gene family has received much attention in Arabidopsis thaliana (Verma and Hong, 2001; Dong et al., 2005; Enns et al., 2005; Evert and Eichhorn, 2006). The 12 Arabidopsis CalS (AtCalS) genes exhibit multiple tissue expression patterns and functions. For example, AtCalS1, AtCalS11 and AtCalS12 are involved in cell plate formation, microspore separation, pollen tube growth, and prevention of pollen degeneration during early development (Hong et al., 2001; Enns et al., 2005). Among AtCalS genes, only AtCalS5 has been implicated in exine development, with low expression in other tissues (Dong et al., 2005; Nishikawa et al., 2005).
Here the results of two complementary lines of enquiry are presented. First, through correlated light, fluorescence and immuno-gold electron microscopy, the occurrence of callose in the moss spore wall, specifically the aperture exine, is demonstrated. The second study is a bioinformatic analysis of the CalS gene family. The report of AtCalS5 affecting pollen exine development in arabidopsis prompted the comparison of the CalS gene family between Physcomitrella and arabidopsis (Preuss et al., 1994; Rhee and Somerville, 1998; Paxon-Sowders et al., 2001; Ariizumi et al., 2003; Dong et al., 2005; Enns et al., 2005; Nishikawa et al., 2005). Annotation of 12 PpCalS genes in P. patens is provided and a phylogenomic analysis of the CalS gene family in moss and Arabidopsis is presented. Specifically, the hypothesis that an exine development orthologue is present in P. patens is tested based on amino acid sequence similarity with AtCalS5.
MATERIALS AND METHODS
Growing Physcomitrella patens
Physcomitrella patens Bruch & Schimp subsp. patens (Ashton and Cove, 1977) protonema was propagated vegetatively in 5·5-cm Petri dishes with BCD medium (Cove, 2000). Gametophores were transferred to sterile 30-mm-diameter peat pellets (Jiffy-7; Jiffy Products International) using tweezers. Pellets were prepared by soaking in distilled water until fully expanded, then autoclaved in Pyrex culture dishes. Water levels were maintained just below the upper surface of the cylindrical pellets. The inoculated peat pellets were cultured at 25 °C under 16-h light/8-h dark conditions for 6 weeks and then transferred to 15 °C under 8-h light/16-h dark conditions to induce gametangia development. Fertilization took place readily in the wet conditions of the peat pellets and capsules at all stages of development were ready for harvest 4 weeks after transfer.
Light microscopy
Capsules were broken open onto glass slides in 0·067 m sodium phosphate buffer, pH 8·5, as a control or stained with 1 % (w/v) aniline blue stain in the same buffer for 1 h each. Aniline blue binds 1,3-β-glucan that emits a yellow-green fluorescence when subjected to UV light. Callose fluorescence was observed using an excitation filter at 350 nm and an emission filter with a transmission cut-off at 460 nm. The slides were viewed on a Leica DM 5000B compound microscope equipped with UV fluorescence and a Q-Imaging Retiga 2000R digital camera.
Immuno-electron microscopy
For immuno-electron microscopy, capsules were fixed in 2·0 % glutaraldehyde in 0·05 m sodium phosphate buffer, pH 7·2, overnight at 4 °C. The fixed capsules were rinsed three times in buffer then serially dehydrated in ethanol. Specimens were slowly infiltrated with Spurr's or LR-White resin (Electron Microscopy Supply) and cured at 60 °C for 48 h (Spurr's) and 7 d (LR-White). The embedded capsules were cut with a diamond ultra knife to achieve sections of 100 nm (i.e. pale gold reflectance). The ultrathin sections were mounted on both 300- and 600-mesh uncoated nickel grids. The grids were placed on a slide warmer (45–50 °C) overnight to stick the sections to the grids. Immunogold preparation was conducted in a humid chamber for the three incubation steps and two washes: 10 µL of 1 % (w/v) bovine serum albumin (BSA) in phosphate-buffered saline (PBS), 30 min; 5 µL of primary monoclonal 1,3-β-glucan antibody (Biosupplies, Australia) diluted 1 : 20 with BSA–PBS, 4 h; followed by three exchanges of BSA–PBS, 5 µL of gold conjugated (size 15 nm) goat-anti-mouse secondary antibody (EMS, USA), 30 min; and four washes of PBS. Grids were then carefully rinsed with nano-pure water to remove residual chloride ions. The grids were air dried at room temperature and stained in 2 % aqueous uranyl acetate for 3 min and in Reynold's lead citrate for 30 s (Ligrone et al., 2002). The grids were examined with a Hitachi 7650 transmission electron microscope and images were collected digitally.
Immuno-light microscopy
Sections of LR-White embedded capsules were cut with a diamond histoknife, mounted on BioBond (EMS, USA)-coated glass slides and labelled using the above procedure. The final staining step was replaced with incubation of the sections in a freshly prepared solution of Amersham IntenSE (Amersham Bioscience, UK) silver enhancement reagent. Negative controls were made for light and electron microscopy by omitting the incubation in the primary antibody.
Genome searches and bioinformatics
Previously identified Arabidopsis thaliana callose synthase sequences (Doblin et al., 2001; Dong et al., 2005, Enns et al., 2005; Nishikawa et al., 2005) were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov). These sequences were used to search for CalS and CalS-like DNA sequences from the Physcomitrella patens ssp. patens (ver 1·1) genome (http://genome.jgi-psf.org/euk_home.html) using BLASTX, BLASTN and BLASTP (low complexity filter on, Blosum62 substitution matrix) (Altschul et al., 1990, 1997). Protein motifs were queried using Pfam (Bateman et al., 2004), PROSITE (Hulo et al., 2006), CDD (Conserved Domain Database) or CDART (Conserved Domain Architecture Retrieval Tool) (Marchler-Bauer and Bryant, 2004; Marchler-Bauer et al., 2005). Domains used for identification were Pfam PF02634 (glucan synthase), CDD 86706 (glucan synthase), IPR003440 (glycosyltransferase) and Pfam 04652 (DU605).
Sequence alignment and phylogenetic analysis
An initial alignment of 24 complete deduced plant CalS amino acid sequences was created in ClustalW (Chenna et al., 2003) using the Gonnet protein weight matrix, pairwise gap opening/extension penalties of 10/0·1 and multiple alignment gap opening/extension penalties of 10/0·5. See Fig. 2 legend for accession numbers. This initial alignment was manually adjusted using GeneDoc (Nicholas et al., 1997). Phylogenetic trees were generated by both neighbor-joining and parsimony methods using PAUP* (version 4.1b10; Sinauer Associates, Sunderland, MA, USA). Support for nodes on the estimated phylogeny was tested with 1000 bootstrap replicates using the parsimony method.
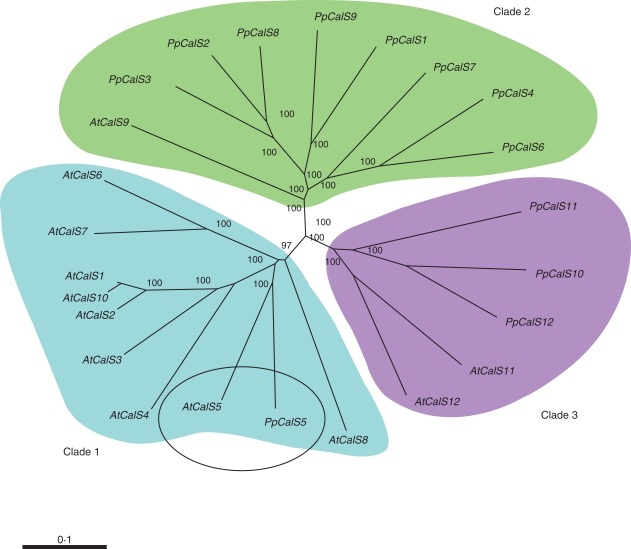
Fig. 2.
Unrooted and bootstrapped gene tree of the CalS gene family from Physcomitrella patens and Arabidopsis thaliana. ClustalW (*) was used to generate an alignment that was manually curated with GeneDoc of the full-length deduced polypeptides. Bootstrap replicates = 10 000 with values reported as percentages for each of the nodes.
RESULTS
General morphology and immunocytochemistry
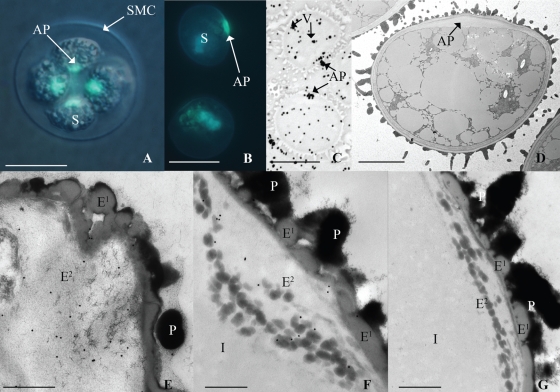
Aniline blue staining of tetrads surrounded by or released from the spore mother cell (SMC) walls revealed a bright yellow-green fluorescence restricted to the proximal side of each developing spore, i.e. the region where the four spores meet in a tetrad. (Fig. 1A, B). This area corresponds to the aperture, a specialized region of the spore wall that permits the protonemata to emerge from the spore coat during germination. In line with the result of aniline-blue staining, silver-enhanced immuno-labelling showed a positive reaction for callose in the aperture; more scattered silver precipitates were also visible in the vacuoles of maturing spores (Fig. 1C). No wall labelling was observed at any other stage of development and controls for all stages were completely negative.
Fig. 1.
Light and electron micrographs of spores from Physcomitrella patens. (A) Spores (S) in a tetrahedral tetrad inside the spore mother cell (SMC) wall stained with aniline blue; positive reaction for callose in aperture (AP) region of spores. (B) Spores liberated from SMC with wall in early developmental stages indicated by lack of spinose morphology stained with aniline blue; positive reaction in the aperture. (C) Positive reaction to silver enhanced anti-callose immunolabel in vacuoles (V) and the aperture of mature spores. (D) TEM of unlabelled spore with expanded aperture. (E–G) TEM of spore wall at the aperture region of proximal wall consists of perine (P), outer exine (E1), inner exine (E2) and intine (I): (E) oblique tangential section through aperture at early developmental stage indicated by relatively perine-free proximal wall has positive reaction to anti-callose immunogold label in the exine; (F) longitudinal section through aperture at later developmental stage indicated by the distinct presence of intine has notable positive reaction to anti-callose immunogold label associated with dark globules in (E2); (G) longitudinal section through aperture labelled with only secondary antibody as a control. Scales bars: (A–C) = 15 µm; (D) = 5 µm; (E–G) = 500 nm.
Observed by electron microscopy in nearly mature spores, the aperture was found to consist of a local expansion of a loosely fibrillar intine layer underlying the exine (Fig. 1D). In the remaining wall the exine is uniform in thickness and consists of two layers, an inner fibrillar layer with scattered electron-opaque globules, and an outer thinner layer of homogeneous lightly opaque material which undulates and in part contributes to spore ornamentation (Fig. 1D–G). An opaque perine is deposited externally on the exine and is responsible for the spinose sculptoderm on the distal spore surface and globules over the proximal surface (Fig. 1D–G). In fully mature spores, perine deposition masks the outer exine layer. Immunogold labelling at the TEM level revealed that callose epitopes were restricted to the inner fibrillar exine layer of the aperture in developing and mature spores. Here the labelling was mainly associated with the dense globules (Fig. 1E and F), whilst virtually no background labelling was observed in other areas of the sections. Control samples labelled with only the secondary gold-conjugated antibody exhibited no labelling anywhere along the spore wall, including the aperture (Fig. 1G).
Sequence alignment and phylogenomic analysis
The assembled plant genome for P. patens (ver 1·1; Rensing et al., 2008) was analysed for the presence of CalS and CalS-like DNA sequences. Physcomitrella genes were named based upon phylogenomic analysis (Fig. 2) and sequence similarity of the deduced polypeptide sequences to previously characterized AtCalS genes from arabidopsis (Table 1) (Verma and Hong, 2001; Dong et al., 2005; Enns et al., 2005; Evert and Eichhorn, 2006). The P. patens genome contains 12 putative callose synthase genes (Table 1). The deduced PpCalS polypeptides range from 1755 to 1966 amino acid residues and are predicted to contain between 11 and 17 transmembrane domains. Nine deduced P. patens polypeptides (PpCalS1–9) have an average size of 1936 amino acids residues (+/– 16·0) and three deduced polypeptides (PpCalS10–12) have an average size of 1761 amino acids residues (+/– 7·0) (Table 1).
Table 1.
Callose synthase (CalS) gene family in the Physcomitrella patens genome (v. 1·1)
| Gene annotation | Accession number | TM domains | Deduced polypeptide |
|---|---|---|---|
| Pp_CalS1 | XP_001772017 | 14 | 1936 |
| Pp_CalS2 | XP_001759994 | 16 | 1930 |
| Pp_CalS3 | XP_001755638 | 17 | 1929 |
| Pp_CalS4 | XP_001764343 | 11 | 1966 |
| Pp_CalS5 | XP_001776005 | 15 | 1930 |
| Pp_CalS6 | XP_001773231 | 12 | 1934 |
| Pp_CalS7 | XP_001766872 | 13 | 1952 |
| Pp_CalS8 | XP_001764315 | 15 | 1942 |
| Pp_CalS9 | XP_001754415 | 12 | 1910 |
| Pp_CalS10 | XP_001783858 | 11 | 1759 |
| Pp_CalS11 | XP_001764182 | 13 | 1755 |
| Pp_CalS12 | XP_001754476 | 13 | 1769 |
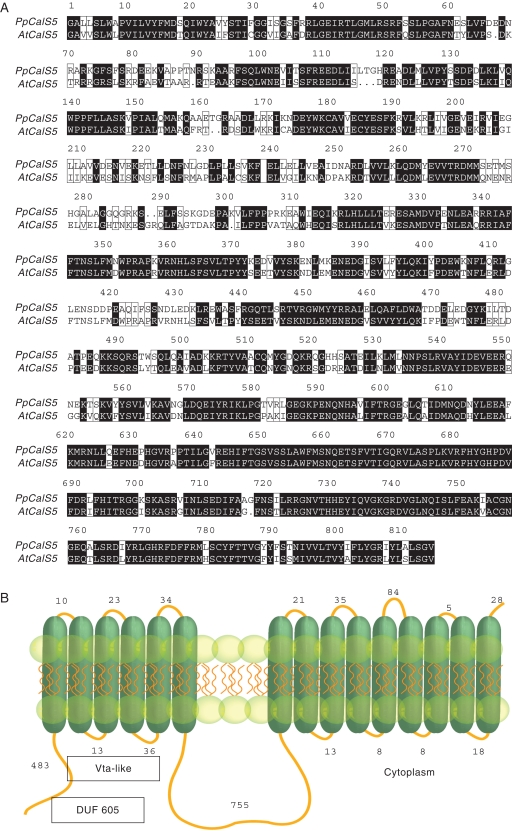
Phylogenomic analysis of the deduced polypeptides from P. patens and A. thaliana strongly supports the presence of three CalS clades (Fig. 2). Clade 1 contains nine A. thaliana sequences (AtCalS1–8 and AtCalS10) and a single P. patens sequence (PpCalS5), Clade 2 contains one A. thaliana sequence (AtCalS9) and eight P. patens sequences (PpCalS1–4 and PpCalS6–9). Clade 3 contains two A. thaliana sequences (AtCalS11,12) and three P. patens sequences (PpCalS10–12). PpCalS10–12 are 170 amino acid residues shorter than PpCalS1–9. PpCalS5 is nested within the AtCalS genes and is least similar to any of the PpCalS genes (Fig. 2). Comparison of the deduced polypeptide PpCalS5 with AtCalS5 reveals that both sequences share a number of protein motifs: DUF605 and Vta-like domains (pfam04652) within the N-terminus and four highly conserved regions in the inner loop (Fig. 3A). Structurally the PpCalS5 consists of a 5' tail 483 amino acids in length, 15 transmembrane domains, and an inner loop 755 amino acids in length (Fig. 3B), while AtCalS5 consists of a 5' tail, 16 transmembrane domains, and an inner loop of similar length.
Fig. 3.
Deduced polypeptide sequence alignment of PpCalS5 and AtCalS5, and predicted membrane topology of PpCalS5. (A) Sequence alignment of inner loop of putative spore exine development callose synthase (PpCalS5) with known pollen exine development callose synthase (AtCalS5). Highly conserved domains shaded black. (B) Predicted protein structure of PpCalS5 with putative conserved DUF 605 and Vta-like domains (pfam04652) in the N-terminus.
DISCUSSION
This is the first report that callose is involved in sporogenesis in mosses. Although callose plays a role in tracheophyte spore and pollen development, liverworts were the only bryophytes conclusively demonstrated to involve callose in spore wall differentiation (Brown and Lemmon, 1987, 1988, 1990). Neidhart (1979) reported the existence of callose in the SMCs of hornworts but this has yet to be verified (Brown and Lemmon, 1990). Callose occurs in meiospore walls of the multicellular green alga Coleochaete (Graham and Taylor, 1986; Brown and Lemmon, 1990; Graham, 1993). In liverworts, callose is involved in pre- (SMC) and post-meiotic exine patterning which leads to the development of elaborate spore surface ornamentation (Brown and Lemmon, 1987). In contrast, in P. patens callose is absent from the SMCs but appears in developing spores and is localized in the expanded aperture region. As spores reach their final stages of development, the aperture collapses and callose is lost. This highly specific localization implies callose is involved in expansion of the aperture during wall development. In seed plants, callose serves to separate the developing microspores and provides a template for pollen exine development (Hong et al., 2001; Dong et al., 2005; Nishikawa et al., 2005). Although this polysaccharide is localized in different spore/pollen wall regions and appears at different developmental stages across plants, the present demonstration of callose in Physcomitrella spores supports the notion that callose involvement in sporogenesis is a signature of embryophytes (Graham and Taylor, 1986; Brown and Lemmon, 1987, 1990; Graham, 1993).
Phylogenomic comparisons between Physcomitrella and Arabidopsis identify three clades of callose synthase genes based on deduced polypeptide sequences. Thus, at least two duplications of the CalS gene occurred prior to the divergence of mosses, some 400 million years ago (Pryer et al., 2004; Newton et al., 2007). Different amino acid sequences may reflect tissue- or development-specific gene expression. Similar interpretations were reported for genes responsible for root hair and rhizoid development (Menand et al., 2007) as well as the cellulose synthase (Roberts and Bushoven, 2007) and β-expansin genes (Carey and Cosgrove, 2007). It is safe to assume that selective pressures specific to the moss versus flowering plant life cycle played a role in CalS gene evolution. Each of the three gene clades contains combinations of genes from both plants, but with varying degrees of gene duplication in the two plants. Like Arabidopsis, Physcomitrella has 12 CalS genes, but with the majority in clade 2. Within this clade a single Arabidopsis gene, AtCalS9 (a.k.a. AtGSL10) shown to affect pollen development at the mitotic stage (Huang et al., 2008), is sister to eight moss genes.
Another intriguing finding is that PpCalS10–12 genes are more similar to each other and to AtCalS11,12 (a.k.a. GSL 1and5) than to any other PpCalS gene. Double mutants of AtCalS11,12 produce smaller flowers, short lateral bolts, shorter roots, and severely deformed pollen with unusual pore structures (Enns et al., 2005). The latter phenotype was the result of failure to form the callose wall that separates the microspores while in the tetrads. Additionally, some of the deformed pollen grains could germinate but were infertile suggesting that AtCalS11,12 genes have partially redundant roles in both the sporophyte and gametophyte (Enns et al., 2005). The functions of orthologs PpCalS10–12 as compared with AtCalS11,12 remain to be determined but may be hypothesized to be expressed in vegetative cell walls. It is interesting to note that the intersporal walls of some liverworts, not mosses, contain callose that separates the spores in tetrads. With the Marchantia genome sequencing project in progress (http://www.jgi.doe.gov/sequencing/cspseqplans2008.html), callose synthase genes for this liverwort may be added soon to the analysis, and will shed light on sequence homology between intersporal wall callose in arabidopsis and liverworts.
With nearly 65 % identity of deduced polypeptide sequences to AtCalS5, the orthologous moss gene PpCalS5 is a likely candidate gene involved in production of the exine callose localized in this study. AtCalS5 has been shown through knockout mutants to exclusively affect pollen exine formation (Dong et al., 2005). In support of this proposed function is that, in tobacco, barley and wheat, callose synthase expression is temporally and spatially correlated to specific tissues and organs (Doblin et al., 2001; Wilson et al., 2006; Voigt et al., 2006). To address this postulate, studies are underway using GFP localization of PpCalS5 in both generations of the life cycle of Physcomitrella.
Conspectus
Sporogenesis is a deeply conserved process and one of the key innovations that played a role in the origin and diversification of land plants (Renzaglia et al., 2000, 2007; Shaw and Renzaglia, 2004; Blackmore, 2007). Although the location and timing of callose during sporogenesis in other plant groups is different from that observed in P. patens, the involvement of this polysaccharide in spore development is widespread in land plants. This raises the question whether similar callose synthase genes operate differently in different lineages of plants, or whether functionally divergent, species-specific callose synthase genes are responsible for differences in callose deposition. The answers to these questions will be elucidated as genomes become available from additional groups of plants.
The moss Physcomitrella patens is an important model system for understanding plant biology and evolution (Cove et al., 1997; Wood et al., 2000; Nishiyama et al., 2003; Frank et al., 2005; Lee et al., 2005; Cove et al., 2006; Quatrano et al., 2007). Physcomitrella has an efficient and well-characterized gene-targeting system (i.e. homologous recombination; Schaefer and Zryd, 1997; Schaefer, 2002). Unparalleled among other model embryophytes, the dominant haploid phase of this moss ensures immediate and direct expression of transformed phenotypes (Lee et al., 2005). An in depth understanding of the biology of P. patens is necessary to understand the genes responsible for transformed phenotypes. This is especially true for complex developmental processes such as spore wall development that require sophisticated labelling techniques and detailed microscopic observation. Indeed, with more plant genomes in the pipeline, the limiting factor in studies of functional and evolutionary genomics may be a lack of knowledge about the plants themselves. Studies, such as the present one, that couple structural development with genomic data, are necessary to generate and answer fundamental questions related to the evolution of development.
ACKNOWLEDGEMENTS
We are grateful for the assistance provided by Integrated Microscopy and Graphics Expertise staff and use of their electron microscope facilities at Southern Illinois University, Carbondale. The authors thank Tammy d'Artenay, Renee Lopez-Smith and Katayoun Mansouri for comments on an earlier version of the manuscript. This project was supported by two NSF Tree of Life initiatives (DEB-0228679, DEB-0531751).
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, et al. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Molecular Biology. 2003;53:107–116. doi: 10.1023/B:PLAN.0000009269.97773.70. [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss Physcomitrella patens. Molecular Genetics. 1977;154:87–95. [Google Scholar]
- Aspinall GO, Kessler G. The structure of callose from the grape vine. Chemistry and Industry. 1957 [Google Scholar]
- Bateman A, Coin L, Durbin R, et al. The Pfam Protein Families Database. Nucleic Acids Research. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S. Pollen and spores: microscopic keys to understanding the earth's biodiversity. Plant Systematics and Evolution. 2007;263:3–12. [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR. Pollen wall development in flowering plants. New Phytologist. 2007;174:483–498. doi: 10.1111/j.1469-8137.2007.02060.x. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. Involvement of callose in determination of exine patterning in three hepatics of the subclass Jungermanniidae. Memoirs of the New York Botanical Garden. 1987;45:111–121. [Google Scholar]
- Brown RC, Lemmon BE. Sporogenesis in bryophytes. Advances in Bryology. 1988;3:159–223. [Google Scholar]
- Brown RC, Lemmon BE. Sporogenesis in bryophytes. In: Blackmore S, Knox RB, editors. Microspores: evolution and ontogeny. New York, NY: Academic Press; 1990. pp. 56–94. [Google Scholar]
- Carey RE, Cosgrove DJ. Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins. Annals of Botany. 2007;99:1131–1141. doi: 10.1093/aob/mcm044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ. The moss, Physcomitrella patens. Journal of Plant Growth Regulation. 2000;19:275–283. [Google Scholar]
- Cove DJ, Bezanilla M, Harries P, Quatrano R. Mosses as model systems for the study of metabolism and development. Annual Review of Plant Biology. 2006;57:497–520. doi: 10.1146/annurev.arplant.57.032905.105338. [DOI] [PubMed] [Google Scholar]
- Cove DJ, Knight CD, Lamparter T. Mosses as model systems. Trends in Plant Science. 1997;2:99–105. [Google Scholar]
- Curatti L, Giarrocco L, Salerno GL. Sucrose synthase and RuBisCo expression is similarly regulated by the nitrogen source in the nitrogen-fixing cyanobacterium Anabaena sp. Planta. 2006;223:891–900. doi: 10.1007/s00425-005-0142-7. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Melis LD, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families. Plant Physiology. 2001;125:2040–2052. doi: 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. The Plant Journal. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and fertility. Plant Molecular Biology. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- Evert RF, Eichhorn SE. Esau's plant anatomy: meristems, cells, and tissues of the plant body: their structure, function, and development. Hoboken, NJ: John Wiley and Sons; 2006. [Google Scholar]
- Frank W, Decker EL, Reski R. Molecular tools to study Physcomitrella patens. Plant Biology. 2005;7:220–227. doi: 10.1055/s-2005-865645. [DOI] [PubMed] [Google Scholar]
- Gabarayeva N, Hemsley AR. Merging concepts: the role of self-assembly in the development of pollen wall structure. Review of Palaeobotany and Palynology. 2006;138:121–139. [Google Scholar]
- Garbary DJ, Renzaglia KS. Bryophyte phylogeny and the evolution of land plants: evidence from development and ultrastructure. In: Bates JW, Ashton NW, Duckett JG, editors. Bryophytes for the Twenty-First Century. Leeds, UK: Maney and British Bryological Society; 1998. pp. 45–63. [Google Scholar]
- Geisler-Lee CJ, Hong Z, Verma DPS. Overexpression of the cell plate-associated dynamin-like GTPase, phragmoplastin, results in the accumulation of callose at the cell plate and arrest of plant growth. Plant Science. 2002;163:33–42. [Google Scholar]
- Gorska-Brylass A. Callose in gametogenesis in liverworts. Bulletin de L'Academie Polonaise des Sciences. 1969;17:549–554. [Google Scholar]
- Graham LE. The origin of land plants. New York, NY: John Wiley and Sons; 1993. [Google Scholar]
- Graham LE, Taylor CE. Occurrence and phylogenetic significance of ‘special walls’ at meiosporogenesis in Coleochaete. American Journal of Botany. 1986;73:597–601. [Google Scholar]
- Graham LE, Delwiche CF, Mishler BD. Phylogenetic connections between the ‘green algae’ and the ‘bryophytes. Advances in Bryology. 1991;4:213–244. [Google Scholar]
- Hong Z, Delauney AJ, Verma DPS. A cell plate-specific callose synthase and its interaction with phragmoplastin. The Plant Cell. 2001;13:755–768. doi: 10.1105/tpc.13.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Chen XY, Rim Y, et al. Arabidopsis glucan synthase-like 10 functions in male gametogenesis. Journal of Plant Physiology. 2008 doi: 10.1016/j.jplph.2008.06.010. doi: 10.1016/jplph.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, et al. The PROSITE database. Nucleic Acids Research. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, et al. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. The Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Knight CD, Knox JP. Physcomitrella patens: a moss system for the study of plant cell walls. Plant Biosystems. 2005;139:16–19. [Google Scholar]
- Ligrone R, Vaughn KC, Renzaglia KS, Knox JP, Duckett JG. Diversity in the distribution of polysaccharide and glycoprotein epitopes in the cell walls of bryophytes: new evidence for the multiple evolution of water-conducting cells. New Phytologist. 2002;156:491–508. doi: 10.1046/j.1469-8137.2002.00538.x. [DOI] [PubMed] [Google Scholar]
- Lugardon B. Pteridophyte sporogenesis: a survey of spore wall ontogeny and fine structure in a polyphyletic plant group. In: Blackmore S, Knox RB, editors. Microspores: evolution and ontogeny. New York, NY: Academic Press; 1990. pp. 95–120. [Google Scholar]
- Machida M, Takechi K, Sato H, et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proceedings National Academy Sciences of the USA. 2006;103:6753–6758. doi: 10.1073/pnas.0510693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Research. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Keke Y, Jouannic S, et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- McIntosh M, Stone BA, Stanisich VA. Curdlan and other bacterial 1,3-β-glucans. Applied Microbiology and Biotechnology. 2005;68:163–173. doi: 10.1007/s00253-005-1959-5. [DOI] [PubMed] [Google Scholar]
- Neidhart HV. Clarke GCS, Duckett JG, editors. Comparative studies of sporogenesis in bryophytes. Bryophyte Systematics. 1979;14:251–280. [Google Scholar]
- Newton AE, Wikström N, Bell N, Forrest LL, Ignatov MS. Dating the diversification of the pleurocarpous mosses. In: Newton AE, Tangney RS, editors. Pleurocarpous mosses: systematics and evolution. Boca Raton, FL: CRC Press; 2007. pp. 337–366. Systematics Association Special Volume Series No. 71. [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS. 1997;4:14. [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology. 2005;5:1471–2229. doi: 10.1186/1471-2229-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin-I T, et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proceedings of the National Academy of Science of the USA. 2003;100:8007–8012. doi: 10.1073/pnas.0932694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxon-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. DEX1, a novel plant protein is required for exine pattern formation during development in Arabidopsis. Plant Physiology. 2001;127:1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Schuettpeltz E, Wolf PG, Schneider H, Smith AR, Cranfill R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. American Journal of Botany. 2004;91:1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Quatrano RS, McDaniel SF, Khandelwal A, Perroud PF, Cove DJ. Physcomitrella patens: mosses enter the genomic age. Current Opinion in Plant Biology. 2007;10:182–189. doi: 10.1016/j.pbi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer A, et al. The genome of the moss Physcomitrella patens reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Renzaglia KS, Duff RJ, Nickrent DL, Garbary DJ. Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny. Philosophical Transactions of the Royal Society London B. 2000;355:769–793. doi: 10.1098/rstb.2000.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia KS, Schuette S, Duff RJ, et al. Bryophyte phylogeny: advancing the molecular and morphological frontiers. The Bryologist. 2007;110:170–213. [Google Scholar]
- Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. The Plant Journal. 1998;15:79–88. doi: 10.1046/j.1365-313x.1998.00183.x. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT. The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Molecular Biology. 2007;63:207–219. doi: 10.1007/s11103-006-9083-1. [DOI] [PubMed] [Google Scholar]
- Schaefer D. A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annual Reviews in Plant Biology. 2002;53:477–501. doi: 10.1146/annurev.arplant.53.100301.135202. [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd J. Efficient gene targeting in the moss Physcomitrella patens. The Plant Journal. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- Scherp P, Grotha R, Kutschera U. Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Reports. 2001;20:143–149. doi: 10.1007/s002990000301. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Renzaglia KS. Phylogeny and diversification of bryophytes. American Journal of Botany. 2004;91:1557–1581. doi: 10.3732/ajb.91.10.1557. [DOI] [PubMed] [Google Scholar]
- Stevenson DW. Ultrastructure of the nacreous leptoids (sieve elements) in the polytrichaceous moss Atrichum undulatum. American Journal of Botany. 1974;61:414–421. [Google Scholar]
- Stone BA, Clarke AE. Chemistry and physiology of higher plant 1,3-β-glucans (callose) In: Stone BA, Clarke AE, editors. Chemistry and biology of (1,3)-β-glucans. Bundoora, Australia: La Trobe University Press; 1992. pp. 365–429. [Google Scholar]
- Verma DPS, Hong Z. Plant callose synthase complexes. Plant Molecular Biology. 2001;47:693–701. doi: 10.1023/a:1013679111111. [DOI] [PubMed] [Google Scholar]
- Voigt CA, Schäfer W, Salomon S. A comprehensive view on organ-specific callose synthesis in wheat (Triticum aestivum L.): glucan synthase-like gene expression, callose synthase activity, callose quantification and deposition. Plant Physiology and Biochemistry. 2006;44:242–247. doi: 10.1016/j.plaphy.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Secco D, Poirier Y. Characterization of the PHO1 gene family and the responses to phosphate deficiency of Physcomitrella patens. Plant Physiology. 2008;146:646–656. doi: 10.1104/pp.107.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Burton RA, Doblin MS, et al. Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta. 2006;224:655–667. doi: 10.1007/s00425-006-0244-x. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Duff RJ. The aldehyde dehydrogenase (ALDH) gene superfamily of the moss Physcomitrella patens and the algae Chlamydomonas reinhardtii and Ostreococcus tauri. The Bryologist. 2009;112 (in press) [Google Scholar]
- Wood AJ, Oliver MO, Cove DJ. Bryophytes as model systems. The Bryologist. 2000;103:128–133. [Google Scholar]