Table I.
Crystallization, Data Collection, and Refinement
| HCoV-229E X-domain | IBV X-domain | IBV X-domain | |
|---|---|---|---|
| Crystallization conditions | 20% PEG 8000 | 30% PEG 4000 | 1.8M (NH4)2SO4 |
| 0.1M Tris | 0.1M Tris | 0.1M Na-citrate pH 5.6 | |
| pH 8.5 | pH 8.5 | 0.2M K-,Na-tartrate | |
| 5% MPD | 0.2M MgCl2 | ||
| Data collection | |||
| Wavelength (Å) | 1.04 | 0.808 | 0.808 |
| Resolution (Å) | 24.22–1.78 (1.88–1.78) | 40.00–1.60 (1.64–1.60) | 31.28–2.10 (2.21–2.10) |
| Space group | P21 | C2221 | P322 |
| Unit-cell parameters | |||
| a (Å) | 33.56 | 42.21 | 78.20 |
| b (Å) | 65.89 | 79.81 | 78.20 |
| c (Å) | 38.02 | 99.74 | 81.70 |
| α (°) | 90 | 90 | 90 |
| β (°) | 110.1 | 90 | 90 |
| γ (°) | 90 | 90 | 120 |
| Solvent content (%, v/v) | 33.73 | 43.12 | 66.88 |
| Overall reflections | 101,730 | 158,270 | 503,497 |
| Unique reflections | 14,479 (1946) | 22,201 (1448) | 17,123 (2231) |
| Multiplicity | 4.2 (3.8) | 7.1 (6.2) | 11.5 (10.4) |
| Completeness (%) | 96.9 (89.9) | 97.9 (97.1) | 98.4 (89.4) |
| Rmergea (%) | 5.2 (15.5) | 10.3 (24.4) | 7.9 (40.1) |
| I/σ(I) | 19.6 (7.8) | 21.7 (6.8) | 25.6 (6.5) |
| Refinement | |||
| Resolution (Å) | 24.22–1.78 | 40.00–1.60 | 31.28–2.10 |
| Rcrystb | 0.165 | 0.169 | 0.199 |
| Rfreeb | 0.205 | 0.203 | 0.231 |
| r.m.s.d. from ideal geometry | |||
| Bonds (Å) | 0.013 | 0.011 | 0.022 |
| Angles (°) | 1.284 | 1.349 | 1.896 |
| Protein atoms | 1283 | 1325 | 1335 |
| Solvent atoms | 146 | 212 | 75 |
| Heteroatoms | 20 | ||
| Ramachandran plot | |||
| Most favored (%) | 91.7 | 92.5 | 92.1 |
| Additionally allowed (%) | 6.9 | 6.8 | 6.6 |
| Generously allowed (%) | 0.7 | 0.0 | 0.7 |
| Disallowed regions (%) | 0.7 | 0.7 | 0.7 |
Values in parentheses are for the highest resolution shell.
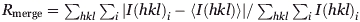 , where I(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over all equivalent reflections.
, where I(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over all equivalent reflections.
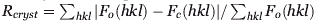 Rfree was calculated for a test set of reflections (6%, 5%, and 6%, respectively) omitted from the refinement.
Rfree was calculated for a test set of reflections (6%, 5%, and 6%, respectively) omitted from the refinement.
