Figure 7.
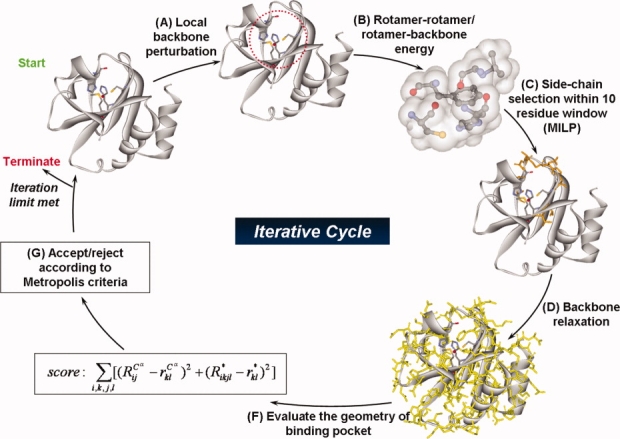
The modified version of protein redesign framework IPRO provides the backbone of the computational environment for the second phase of this study. During each iteration, a local backbone perturbation window (i.e., 1–5 residues) is randomly selected, and a perturbation of the backbone is imposed. An optimization step is carried out globally using an MILP model within a local perturbation window to identify new residues (i.e., mutations) and corresponding rotamers. This optimization step is followed by backbone relaxation. If the redesign and corresponding structural modifications lead to an improved energy of the system while maintaining and/or lowering the overall geometry score, then the perturbation is accepted; otherwise, a simulated annealing step is used to accept or reject the residue redesigns associated with each backbone perturbation step. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
