Figure 3.
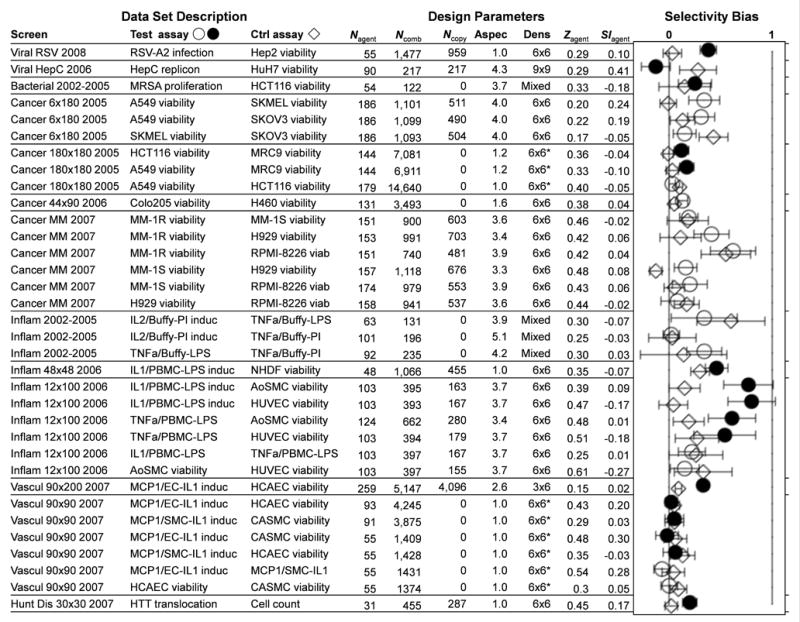
Selectivity bias for thirteen sets of combination data focused on six disease areas. The table shows the “test” and “control” assays corresponding to each comparison, the number of single agents Nagent or combinations Ncomb tested, the number Ncopy of combinations with independent replicates, and the aspect ratio “Aspec” of the agent lists that were combined. Also shown are the sampling “Dens” (dose matrix size, * when sparse), with the average activity Zagent and selectivity index SIagent across the single agents. For each screen, all pairs of assays were compared, in “forward” (circles, filled when aligned with a therapeutic objective) and “reverse” (diamonds) order relative to the assay designations listed. Each combination’s SI value was calculated at Zcut = max(Ztest)/2, and the top 5% of synergies for each test assay were used to determine a selectivity bias. Error bars represent 95% confidence with a sequential multiple hypothesis adjustment48 to account for all assay comparisons in each screen. Weighted by these errors, the consensus selectivity bias is 0.104±0.010 (0.214±0.021 for therapeutically aligned pairs). The synergistic combinations have statistically more positive selectivity, with some screens showing more than threefold potency shifts.
