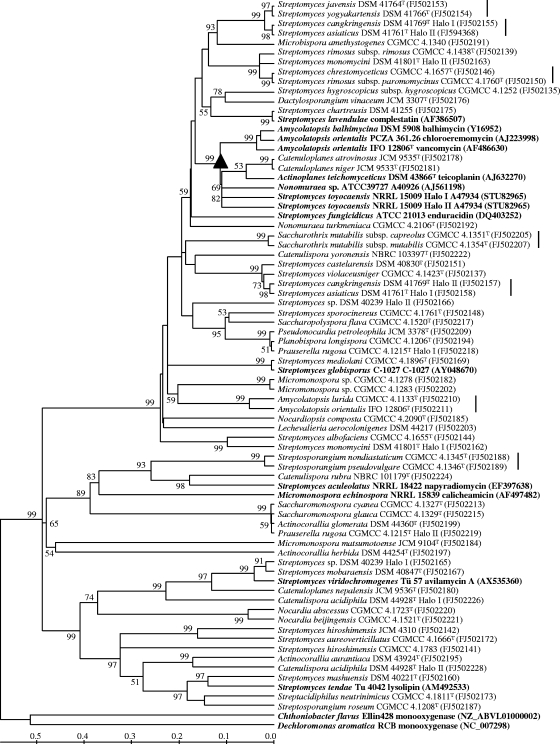
FIG. 1.
Neighbor-joining tree based on halogenase amino acid sequences of 54 reference strains and known halometabolite-producing strains (in boldface). Two closely related flavin-dependent monooxygenase sequences are used as outgroups (in boldface). GenBank accession numbers are given in parentheses. The clade of vancomycin group antibiotics is marked with a triangle. Strains containing multiple halogenase genes are differentiated by the terms Halo I and Halo II. Taxonomically closely related strains sharing highly homologous halogenases are marked with vertical lines to the right of the strain names. Percentage bootstrap values based on 1,000 resampled datasets are shown at the nodes; only values above 50% are given. The scale bar indicates the amino acid substitution rate.