Abstract
We developed a real-time quantitative PCR (qPCR) assay targeting the rRNA internal transcribed spacer region of the hard clam pathogen QPX. The qPCR assay was more sensitive than was histology in detecting clams with light QPX infections. QPX was detected in 4 of 43 sediment samples but in none of 40 seawater samples.
The thraustochytrid called QPX (for quahog parasite unknown) has caused high mortalities in hatchery-reared and wild hard clams (Mercenaria mercenaria, also known as quahogs) from Prince Edward Island (Canada) to Virginia (United States) since the late 1950s (17, 22, 25, 29). In the summer of 2002, QPX infections appeared in the previously healthy Raritan Bay (off the coast of Staten Island in New York) M. mercenaria population, causing significant clam mortality and closure of the fishery (6). Management of hard clam populations affected by QPX disease is hampered by an incomplete understanding of factors controlling the occurrence and severity of QPX infections. Environmental factors, such as salinity and temperature, appear to be important (22), as do clam population density and the planting of seed from nonlocal sources (7). More quantitative information about the occurrence and progression of QPX disease in relation to these and other variables would support better prediction of, and response to, QPX outbreaks. QPX is thought to be an opportunistic pathogen (4, 7, 11), capable of growing outside its host. However, there is very little known about substrates that might support QPX organisms outside of hard clams (4). The abilities to detect and enumerate QPX cells in potential reservoirs would allow the dynamics of the QPX organism in the environment to be related to the occurrence of QPX disease, offering new insight into fundamental questions about the natural transmission mechanisms of the infection.
The 18S ribosomal DNA (rDNA) primer pair QPX-F and QPX-R2 can be used in a standard PCR assay to detect the presence of QPX DNA in clam tissue samples (26). Unfortunately, the products are too long (∼650 bp), and often include too much primer dimer, for use in a SYBR green real-time quantitative PCR (qPCR) assay. The low sequence variability in rRNA genes made it difficult to design other primers specific for QPX 18S rDNA. Instead, we used our previously reported rRNA internal transcribed spacer (ITS) region (including ITS1, the 5.8S rRNA gene, and ITS2) sequences for QPX isolates from Massachusetts and New York (20) to develop a qPCR assay targeting the more variable ITS region (1).
Development of QPX-specific real-time qPCR assay.
The ITS regions of the thraustochytrids Schizochytrium aggregatum (ATCC 28209), Schizochytrium limacinum (ATCC MYA-1381), and Thraustochytrium aureum (ATCC 34304) (acquired from the American Type Culture Collection, Manassas, VA, and maintained in medium 790 By+ at 23°C) were PCR amplified with universal 18S and 28S rDNA primers (18S-RR and 28S46Rev) (Table 1), cloned and sequenced as described previously (20), and submitted to GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers FJ533155 to FJ533163. These sequences were aligned (using BioEdit version 7 [13]) in the 5.8S rRNA gene and, where possible, in ITS1 and ITS2, with ITS region sequences from QPX (GenBank accession numbers DQ641197 to DQ641141 [20]), three Aplanochytrium strains (labyrinthulids more distantly related to QPX; GenBank accession numbers EU872090 to EU872092), and more than 30 species representing 12 major groups of heterokonts. The 5.8S rRNA genes of QPX and the other labyrinthulids were between 93.3% and 88.7% identical, while their ITS1 and ITS2 region sequences were so different that they could not be aligned (data not shown). Primer 5.8S24For (Table 1) was designed to match QPX 5.8S rDNA, and it mismatched the other thraustochytrids at one or two bases. Primer QPX-ITS2-R2 (Table 1) was designed with one degenerate base to match all known variants of the QPX ITS2 sequence (20), and it did not match any other sequences in the alignment at more than a few bases. The melting temperatures and secondary structures of the primers were analyzed with Primer Premier 5 software (Premier Biosoft Inc., Palo Alto, CA). BLAST against GenBank revealed that while each of the 5.8S24For and QPX-ITS2-R2 primers does match sequences from a variety of other organisms (or mismatches at only one or two bases), only the QPX ITS region sequences match both primers.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′→3′) | Target | Application(s) | Reference or source |
|---|---|---|---|---|
| QPX-F | ATCCTCGGCCTGCTTTTAGTAG | QPX (18S rDNA) | PCR | 26 |
| QPX-R2 | GAAGTCTCTACCTTTCTTGCGA | QPX (18S rDNA) | PCR | 26 |
| Laby-A | GGGATCGAAGATGATTAG | Labyrinthulids (18S rDNA) | PCR | 26 |
| Laby-Y | CWCRAACTTCCTTCCGGT | Labyrinthulids (18S rDNA) | PCR | 26 |
| 18S-RR | GTAGGTGAACCTGCAGAAGGATCA | Eukaryotes (18S rDNA) | PCR and cloning | 18 |
| 5.8S24For | TTTAGCGATGGATGTCT | QPX (5.8S rDNA) | PCR and qPCR | This study |
| QPX-ITS2-R2 | GCCCACAAACTGCTCTWT | QPX (ITS2 region) | PCR and qPCR | This study |
| 28S46Rev | ACCCGCTGAARTTAAGCATAT | Eukaryotes (28S rDNA) | PCR and cloning | 27 |
Primers were synthesized by Integrated DNA Technologies (Coralville, IA) and tested under standard PCR conditions (95°C for 30 s, 55°C for 1 min, and 72°C for 2 min for 35 cycles; then 72°C for 10 min) in 25-μl reaction mixtures containing 1× PCR MasterMix (Eppendorf Inc., Westbury, NY), 200 nM of each primer, and 1 μl of template DNA. The 5.8S24For and QPX-ITS2-R2 primer set produced products of the expected size without artifacts (nonspecific amplification products or primer dimers) for all QPX-positive templates tested, including plasmids carrying cloned QPX ITS region DNA, purified DNA of cultured QPX cells, and DNA isolated from QPX-infected clams. No amplification products were generated with the QPX-negative templates (no DNA control, genomic DNA from three other thraustochytrids, and DNA from clam tissue that was diagnosed as QPX-free by histology and by PCR with the QPX-F and QPX-R2 primers) (Table 1). Additionally, the 5.8S24For and QPX-ITS2-R2 primer set gave no amplification products with template DNA purified from QPX-free (as determined by the QPX-F and QPX-R2 primers) seawater samples, even though the more general labyrinthulid 18S rDNA primer set, Laby-A and Laby-Y (26), gave abundant PCR products, confirming that labyrinthulids other than QPX were present in the samples but were not amplified by the QPX-specific primers.
qPCR assays were done with a Stratagene (La Jolla, CA) MX3000P thermal cycler and software (version 2.0). Each 25-μl reaction mixture contained 1× core PCR buffer, 2 mM MgCl2, 0.8 mM deoxynucleoside triphosphate mix, 8% glycerol solution, 3% dimethyl sulfoxide, 30 nM reference dye, 0.167× SYBR green I dye, and 1.25 U SureStart Taq DNA polymerase from the Stratagene brilliant SYBR green qPCR core reagent kit, 100 nM each primer, and 1 μl DNA template. The real-time PCR program was 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 1 min at 55°C, and 1 min at 72°C. Individual well fluorescence data were collected at the end of each cycle, and the fluorescence detection threshold was determined automatically. The signal from QPX-negative templates did not cross the threshold within 40 cycles. Thermal dissociation curve analysis performed at the end of each run showed that the products amplified from QPX-positive templates had a single peak at 80 to 81°C. Replicate reactions generally agreed very well, with the standard deviation of the mean CT value (the cycle number at which SYBR green fluorescence crossed the detection threshold) being typically less than 0.2. The average CT value of the two or three replicate reactions for each template was used for further data analysis.
The standard curve relating the initial QPX ITS region copy number to the CT value was generated by 10-fold serial dilution, ranging from 10 to 106 copies, of plasmids containing the QPX ITS region amplified by primers QPX-F and 28S46Rev (Table 1). Plasmid DNA was purified by using the Wizard Plus SV miniprep DNA purification system (Promega, Madison, WI) according to the manufacturer's instructions. The concentration of plasmid DNA was measured fluorometrically with a PicoGreen double-stranded DNA quantification kit (Molecular Probes, Eugene, OR) and a TBS-380 mini fluorometer (Turner Biosystems, Sunnyvale, CA) following the manufacturer's instructions. To reflect the known variation in QPX ITS region sequences (20), the plasmid standard was a mixture of three clones (GenBank accession numbers DQ641187, DQ641192, and DQ641195). The differences in length (188 versus 189 bp) and sequence (2 bp) among qPCR products from the three plasmids were not detectable in thermal dissociation curves of the standards. Averaged over 35 independent qPCR assays with this standard curve (run in duplicate or triplicate each time), the 5.8S24For and QPX-ITS2-R2 primer set gave a strong linear (R2 = 0.99) inverse relationship between the CT value and the log10 initial copy number of plasmid, with a slope of −3.84 ± 0.21 (mean ± standard deviation), and was capable of detecting as few as 10 QPX ITS copies per reaction, which corresponded to a mean CT value of 36.96 ± 1.68.
The DNA content of QPX cells was determined by pooling 8 to 10 QPX isolates growing in the exponential phase, removing the mucoid material enveloping the cells as reported previously (20), and counting the cells in a hemocytometer (Hausser Scientific, Horsham, PA) at ×400 magnification until the total volume of counted cell suspension reached 1.5 to 2 μl. DNA was extracted from 1.0 × 106 QPX cells by the method of Galluzzi et al. (9) and quantified by PicoGreen, giving an estimate of 257.1 ± 37.4 fg DNA cell−1 (the grand mean of triplicate subsamples from three independent sets of counted QPX cells). Based on this value, DNA purified from cultured QPX cells with the BD NucleoSpin tissue kit (BD Biosciences, Macery-Nagel, Germany) was used to set up a series of reactions with DNA equivalent to 0.5, 1, 5, 50, 500, and 5,000 QPX cells. The number of ITS copies in each of these reactions was calculated by plotting the CT value on the plasmid standard curve, giving an estimate of 181 ± 68 ITS copies in each QPX cell (the grand mean of 10 different determinations). This value is within the range reported for other heterokonts of similar sizes (19, 31). With a conservative detection limit of 10 ITS region copies per reaction, our QPX qPCR assay can detect approximately 0.05 QPX cells per reaction, substantially lower than the 1 cell per reaction detection limit of the protein-coding gene qPCR assay for QPX developed by Lyons et al. (15).
A potential complication in estimating the abundance of QPX cells by any molecular genetic method arises from its life history (14). A single thallus in the process of becoming a sporangium could have dozens of copies of the genome but be counted microscopically as a single cell. Because the proportion of thalli and sporangia varies with the growth phase of a culture (Q. Liu, unpublished data), the genomic DNA content and average number of ITS copies per cell would be expected to vary accordingly. Our estimated genomic DNA content would indicate a genome size of approximately 251 Mbp for QPX, which is 20- to 25-fold greater than the genome sizes recently estimated for four other thraustochytrids (2). Further work will be required to determine whether the apparently high DNA content of QPX reflects its life history or other factors. We assumed that the rDNA copy number of QPX cells in culture is the same as that of QPX cells in field samples, and we used the estimated number of ITS region copies per QPX cell to convert the detected ITS region copy number in clam and environmental samples to cell counts (see below). The life stages of QPX in culture and in infected clams appear very similar (14), so estimates of QPX cell numbers based on cultured cells should be reasonable for at least clam tissue samples.
Sample collection and processing.
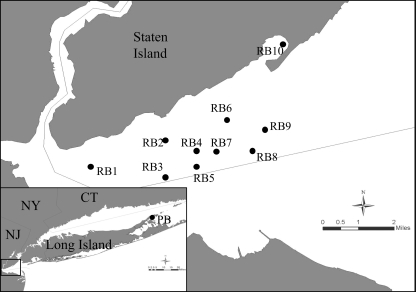
Clam, seawater, and sediment samples were collected from 10 sites in Raritan Bay plus one site in Hashamomuck Pond in Peconic Bay (both in New York) (Fig. 1) during five visits at 6-week intervals from April to September 2006. At each sampling site, 4 liters of seawater from 1 meter below surface was collected with a 2-liter General Oceanics (Miami, FL) Niskin bottle, and 500 ml was filtered through a Sterivex-GV filter unit (Millipore, Billerica, MA) with a peristaltic pump. Sediment samples were collected from the patent tong, which was also used to collect clam samples (30 clams at each site), by pooling and mixing the intact surface layer (0.5 to 1 cm deep) in a sterile container and then transferring subsamples to 2-ml cryovials. The filters and sediment samples were stored immediately on dry ice and transferred to a −80°C freezer until DNA extraction. After measurement of shell length and width and gross examination for abnormal signs, such as nodules or swelling, each clam was dissected. A thin cross section of clam meat, containing mantle, siphon, gills, digestive glands, stomach, gonad, pericardium, and kidney, was fixed in 10% buffered formalin and processed for histological examination (6). The remaining mantle and siphon tissue was weighed and preserved in 100% ethanol at −80°C until DNA extraction.
FIG. 1.
Locations of the Hashamomuck Pond (Peconic Bay [PB]) and Raritan Bay (RB) sampling sites at the east end of Long Island and south of Staten Island, NY. Samples were not collected from RB5 or RB6 in April; from RB5, RB6, or RB8 in May; or from RB3, RB5, or RB6 in June or in September. The sediment sample from RB8 in June was lost, as were the seawater samples from RB3 in April and May and from RB5 and RB6 in August.
qPCR assay of QPX in clam tissues.
QPX infections are typically microscopic and focal or multifocal (6, 22, 25) and can be missed if only a small amount of tissue is examined. To minimize the risk of false-negative qPCR results, we washed the ethanol-preserved clam tissues twice in phosphate-buffered saline, mechanically homogenized the whole mantle and siphon (excluding the thin section used for histology) in 10 volumes of phosphate-buffered saline, and for routine assays, extracted DNA from a 1-ml aliquot containing 100 mg clam tissue. Cells were harvested from the homogenate by centrifugation at 12,000 × g for 10 min and resuspended in 250 μl T1 lysis buffer and 25 μl proteinase K (20 mg ml−1) from the BD NucleoSpin tissue kit, incubated at 56°C overnight (16 to 18 h), and processed further by following the manufacturer's protocol. The column was eluted twice with 75 μl of preheated elution buffer, and the two eluates were combined. To determine the recovery rate of QPX DNA by this procedure, DNA was extracted side by side from aliquots of clam tissue homogenate spiked with 1.0 × 105 QPX cells and aliquots to which no QPX cells were added. The recovery rate (percent) of DNA from the spiked QPX cells was calculated with the following formula: (QPXrecovered/QPXadded) × 100, where QPXrecovered is the difference between the numbers of QPX ITS copies measured in the “spiked” and “unspiked” samples divided by 181 ITS copies per QPX cell and QPXadded is the number of QPX cells added. Using only 25 mg of clam tissue homogenate rather than 100 mg may have improved the recovery of QPX DNA twofold, although the difference was not statistically significant. The recovery rate did differ significantly between two of the NucleoSpin tissue kits used during this study—0.9 % ± 0.5% (n = 3) for one and 16.3% ± 12.3% (n = 3) for the other. The recovery of DNA from QPX cultures using the same two kits (estimated by comparison to the QPX DNA content measured by the Galluzzi et al. (9) method described above also differed significantly—4.7% ± 0.6% (n = 3) for one kit and 26.4% ± 2.7% (n = 3) for the other. A third NucleoSpin tissue kit, which was not used for clams in this study, gave a DNA recovery rate from QPX cultures that was statistically indistinguishable from the second kit (36.6% ± 8.9% [n = 3]). Since we used the same protocols, instruments, and personnel throughout, the difference between kits was most likely due to differences in the kit reagents, and this suggests that the recovery rates of extraction procedures should be estimated frequently, especially when any element of the extraction procedure is changed.
One major difficulty in examining environmental samples by qPCR is the possibility of false-negative results or underestimates of target gene abundance caused by PCR inhibitors that are coextracted with DNA and can vary from sample to sample (3, 5, 8, 9, 12, 24, 28, 30). To measure PCR inhibition in the environmental DNA templates, additional “inhibition control” reactions for each sample were run by adding 1.0 × 103 copies of the same plasmids used in the standard curve. In the regular duplicate or triplicate qPCR assay reactions for each DNA template, QPXtemplate × (100% − I) = QPXassay, and in the inhibition control reactions for each template, (QPXtemplate + QPXadded) × (100% − I) = QPXcontrol, where I is percent inhibition, QPXtemplate is the number of QPX ITS copies in the template, the value of QPXadded is known (1.0 × 103 copies), and the values of QPXassay and QPXcontrol are determined by comparing the assay and inhibition control CT values, respectively, with a plasmid standard curve run in the same plate. The two unknown values, I and QPXtemplate, can be calculated as I = [1 − (QPXcontrol − QPXassay)/QPXadded] × 100% and QPXtemplate = QPXassay/(100% − I). This approach assumes that amplification of the added plasmids is inhibited in the same way as amplification of the target gene purified from the field sample and that inhibitors extracted from the field sample interfere with PCR amplification to the same degree in the parallel assay and inhibition control reactions. For clam tissue samples extracted with the BD NucleoSpin tissue kit, 56 of 74 templates had less than 50% PCR inhibition (average I for those 56 was 13.4% ± 20%), and 10-fold dilution reduced PCR inhibition to less than 50% in 14 of the 18 remaining templates (average I for those 14 was 4.5% ± 23.7%). When I was less than 50%, the qPCR assays were accepted as effective PCR amplifications, and the value of I was used to calculate QPXtemplate. When I was greater than 50%, the template was diluted and assayed again until an effective amplification was achieved.
The original abundance of QPX cells in each clam sample was computed as (QPXtemplate × a × b)/(c × d × e), where a is the dilution factor of the DNA template, b is the total elution volume of the DNA template (usually 150 μl), c is the ITS region copy number per QPX cell (181 cell−1), d is the DNA recovery rate of the extraction method, and e is the weight of clam tissue (usually 100 mg) from which DNA was extracted. To compare the qPCR assay with the histological diagnosis method, we analyzed two groups of clams (30 clams per group) collected in August 2006 from two Raritan Bay sites (RB9 and RB7) where clams have been experiencing QPX disease. Four clams from RB9 and none from RB7 were diagnosed QPX positive by histology (Table 2). QPX was above the qPCR detection limit (0.9 ± 0.4 cells mg tissue−1) in all four histology-positive clams (Table 2). QPX was also above the qPCR detection limit in seven other clams from RB9 and in three clams from RB7. In 8 of the 10 histology-negative but qPCR-positive clams, the estimated QPX abundance was less than 20 cells mg tissue−1. Like the standard PCR assay developed previously (26), the qPCR assay is a more sensitive diagnostic tool especially when relatively few QPX cells are present.
TABLE 2.
QPX abundance in clams from two sites in Raritan Bay in August 2006 diagnosed by histology and qPCR assayd
| Site | Clam no. | No. of QPX ITS copies per reactiona | No. of QPX cells mg tissue−1b | Histology diagnosisc |
|---|---|---|---|---|
| RB9 | 3 | 269 | 13.7 | − |
| 7 | 51,432 | 2,613.3 | + | |
| 11 | 20 | 1.0 | − | |
| 14 | 2,461 | 125.1 | + | |
| 17 | 104 | 5.3 | − | |
| 18 | 3,772 | 191.7 | − | |
| 19 | 1,936 | 98.4 | + | |
| 24 | 30 | 1.5 | − | |
| 26 | 16 | 0.8 | − | |
| 27 | 144 | 7.3 | − | |
| 30 | 5,721 | 290.7 | + | |
| RB7 | 17 | 30 | 1.5 | − |
| 23 | 5115 | 259.9 | − | |
| 26 | 36 | 1.8 | − |
QPXtemplate.
Calculated with the following values: a = 1, b = 150 μl, c = 181 copies cell−1, d = 16.31%, and e = 100 mg.
Diagnosis of QPX disease by histology. +, positive; −, negative.
Prevalence of QPX by qPCR is 36.7% for RB9 clams and 10% for RB7 clams; prevalence of QPX disease by histology is 13.3% for RB9 clams and 0% for RB7 clams.
qPCR assay of QPX in seawater.
The recovery of QPX DNA from seawater was estimated by spiking 2.0 × 105 QPX cells into at least three replicates of randomly selected seawater samples for each of the six extraction methods tested. QPX DNA recovery was 2.9% ± 2.4% (n = 3) with the BD NucleoSpin tissue kit, 1.3% ± 0.1% (n = 3) with the Qiagen (Valencia, CA) DNeasy tissue kit, 4.0% ± 0.4% (n = 3) with the BD NucleoSpin plant kit, and 4.7% ± 1.4% (n = 3) with the BD NucleoSpin plant kit lysis buffer combined with the FastDNA Spin kit for soil (Qbiogene, Carlsbad, CA); differences among these methods were not statistically significant. Recovery with the Qiagen DNeasy tissue kit was improved significantly (to 5.0% ± 1.1% [n = 3]; P < 0.05) when its lysis buffer was replaced with Galluzzi's crude lysis buffer (9). Combining Galluzzi's crude lysis buffer with the physical lysis (bead-beating) method of the FastDNA Spin kit for soil provided still higher recovery (though the difference was not statistically significant) of QPX DNA (9.5% ± 8.2% [n = 9]). For routine sample analysis, DNA was extracted from the seawater particulate matter collected on Sterivex filters by adding 1 ml of freshly made Galluzzi's crude lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.5% Nonidet P-40, 0.5% Tween 20, and 0.1 mg ml−1 proteinase K) (9) to the thawed filter units and incubating at 60°C for 3 h in a rotisserie oven. A 500-μl portion of lysis mixture was transferred to a lysing matrix E tube from the FastDNA Spin kit, and the sample was processed further by following the manufacturer's instructions. DNA was eluted twice in 75 μl DNA Elution Solution, and the two eluates were combined.
In preliminary experiments, all seawater DNA templates from all tested extraction methods had greater than 50% PCR inhibition, and 1:100 dilution was required to reduce inhibition to less than 50%. After treatment of extracted seawater DNA with the StrataPrep PCR purification kit (Stratagene, La Jolla, CA), all templates were still more than 50% inhibited, but 1:10 dilution reduced inhibition to less than 50% in most (48 of 56; 3.9% ± 28.4% average I), and so routine analyses used 1:10 dilutions of DNA treated with the StrataPrep kit. QPX was below the detection limit of 4.2 ± 1.7 cells ml−1 in all 40 Raritan Bay and Hashamomuck Pond seawater samples assayed. This could reflect a low abundance of QPX in seawater combined with low recovery of QPX DNA from seawater samples. Better sensitivity might be achieved by collecting particulate matter on filters with a larger pore size than the 0.22-μm-pore-size filters used here; for example, a 1-μm-pore-size filter would still capture QPX cells (which typically range from 2 to 20 μm in diameter [14]) while allowing larger volumes of water to be filtered and reducing the contribution of bacteria to the extracted DNA. Alternatively, if QPX is associated mainly with marine aggregates, where it has previously been detected (15, 16), much larger particles might need to be collected from larger volumes of seawater in order to routinely detect QPX. The efficiency of QPX cell lysis may have been a factor limiting the recovery of QPX DNA. For example, both the T1 lysis buffer from the NucleoSpin tissue kit and a sodium dodecyl sulfate-based lysis buffer (1% sodium dodecyl sulfate, 25 mM EDTA, 0.1 mg ml−1 proteinase K), even when combined with sonication, yielded less than 10% as much DNA from counted samples of QPX cells than the Galluzzi et al. (9) lysis buffer, which contains Tween 20 and Nonidet P-40 (Q. Liu, unpublished).
qPCR assay of QPX in sediment.
DNA was extracted from 500 to 850 mg (wet weight) of thawed sediment by using the FastDNA Spin kit for soil and following the manufacturer's instructions. The column was eluted twice with 75 μl DNA Elution Solution, and the two eluates were combined. Extracted sediment DNA was treated with the StrataPrep PCR purification kit, which reduced PCR inhibition enough that nearly half of the sediment templates assayed without dilution (5 of 11) and most templates assayed with 1:10 dilution (38 of 49) had less than 50% inhibition (average I for the 38 templates was 3.3% ± 24.7%), so 1:10 dilution of StrataPrep-treated DNA was used routinely for the initial assay of each sediment DNA template. The recovery of QPX DNA by this method from randomly selected sediment samples spiked with 2.0 × 105 QPX cells was 5.4% ± 2.1% (n = 6). The QPX level was below the detection limit of 5.6 ± 2.1 cells mg sediment−1 in 39 of 43 Raritan Bay and Hashamomuck Pond sediment samples, although it was detected at site RB4 (25 ITS copies per reaction, 34 QPX cells mg sediment−1) and site RB9 (474 ITS copies per reaction, 169 QPX cells mg sediment−1) in May, as well as at site RB7 (84 ITS copies per reaction, 52 QPX cells mg sediment−1) and site RB8 (337 ITS copies per reaction, 215 QPX cells mg sediment−1) in September. Two of these templates (RB9 May and RB8 September) were amplified by standard PCR with the qPCR primers for a total of 65 cycles to generate enough products for cloning, and the sequences of four cloned PCR products matched perfectly one or more of the QPX ITS region sequences in GenBank (data not shown).
Thraustochytrids are abundant in coastal benthic habitats (21), and QPX has previously been detected in hard clam pseudofeces (15, 16) and in sediment samples (11). The natural transmission mechanism of QPX disease is not yet known, but sediment, as the habitat of clams and a potential environmental reservoir for QPX, could play a role. The four QPX-positive Raritan Bay samples reported here provide the first estimates of the abundance of QPX in sediments and were collected from four different sites where QPX disease has been occurring in clams since 2002 (B. Allam, unpublished data), suggesting that the presence of QPX in sediment may be related to QPX disease in local clams. However, QPX was detected in too few sediment samples to offer any insight into relationships between QPX abundance in sediment and QPX disease in clams. One limitation of the methods we employed is that in small samples from heterogeneous environments such as sediments, estimates of abundance could be much greater than the average in some places but much lower than that in others. Increasing the amount of sediment processed for each DNA extraction is likely to overload the binding column and increase PCR inhibition. However, homogenizing a much larger sample prior to removing subsamples, as for the clam tissue analysis, or analyzing several replicate samples from one field site may improve the assay. While the FastDNA Spin kit for soil gives a high yield of DNA from soil and sediment samples (5, 23, 24), modifications may be needed to improve recovery of QPX DNA.
To summarize, we have developed a specific and sensitive real-time qPCR assay for QPX with primers targeting the rRNA ITS region. The qPCR assay revealed the presence of relatively small numbers of QPX cells in many clams that were diagnosed disease-free by the histological method and provided the first quantitative evidence for the presence of QPX in sediments associated with an outbreak of QPX disease. Quantification of PCR inhibition allowed us to identify false-negative results and to correct estimates of QPX abundance in samples that gave positive results. Sample processing and DNA extraction and purification methods critically influenced the detection limit of the qPCR assay, which varied among samples of each type depending on PCR inhibition. Gast et al. (10) developed a denaturing gradient gel electrophoresis assay for QPX, using nested PCR that has lower detection limits than this qPCR assay, but the denaturing gradient gel electrophoresis assay is not quantitative. Improved recovery of QPX DNA and/or removal of PCR inhibitors would improve the qPCR assay detection limit. Real-time qPCR offers a promising tool for describing the distribution and dynamics of QPX in the marine environment and for investigating relationships between QPX in the environment and the development of QPX disease within affected clam populations.
Acknowledgments
This research resulted from projects R/XG-15 and R/FBM-33 funded by awards NA16RG1645 and NA07OAR4170010, respectively, from the National Sea Grant College Program of the U.S. Department of Commerce's National Oceanic and Atmospheric Administration, to the Research Foundation of the State University of New York on behalf of the New York Sea Grant. The qPCR assays were performed in the Molecular Evolution of Adaptation and Diversity (MEAD) lab, founded at Stony Brook University with funding from the National Science Foundation (DBI-0400829) to J.L.C. and John True.
The statements, findings, conclusions, views, and recommendations presented in this work are those of the authors and do not necessarily reflect the views of any of those organizations.
We thank the New York State Department of Environmental Conservation for field support and S. Pawagi for help with sample processing.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Alvarez, I., and J. F. Wendel. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29:417-434. [DOI] [PubMed] [Google Scholar]
- 2.Anbu, P., D. U. Kim, E. J. Jeh, Y. S. Jeong, and B. K. Hur. 2007. Investigation of the physiological properties and synthesis of PUFAs from thraustochytrids and its electrophoretic karyotypes. Biotechnol. Bioprocess Eng. 12:720-729. [Google Scholar]
- 3.Audemard, C., L. M. Ragone Calvo, K. T. Paynter, K. S. Reece, and E. M. Burreson. 2006. Real-time PCR investigation of parasite ecology: in situ determination of oyster parasite Perkinsus marinus transmission dynamics in lower Chesapeake Bay. Parasitology 132:827-842. [DOI] [PubMed] [Google Scholar]
- 4.Buggè, D. M., and B. Allam. 2007. Effects of starvation and macroalgae extracts on the survival and growth of quahog parasite unknown (QPX). J. Exp. Mar. Biol. Ecol. 348:60-69. [Google Scholar]
- 5.Cook, K. L., and J. S. Britt. 2007. Optimization of methods for detecting Mycobacterium avium subsp. paratuberculosis in environmental samples using quantitative, real-time PCR. J. Microbiol. Methods 69:154-160. [DOI] [PubMed] [Google Scholar]
- 6.Dove, A. D. M., P. R. Bowser, and R. M. Cerrato. 2004. Histological analysis of an outbreak of QPX disease in hard clams Mercenaria mercenaria in New York. J. Aquat. Anim. Health 16:246-250. [Google Scholar]
- 7.Ford, S. E., J. N. Kraeuter, R. D. Barber, and G. Mathis. 2002. Aquaculture-associated factors in QPX disease of hard clams: density and seed source. Aquaculture 208:23-38. [Google Scholar]
- 8.Frostegård, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garces, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gast, R. J., E. Cushman, D. M. Moran, K. R. Uhlinger, D. Leavitt, and R. Smolowitz. 2006. DGGE-based detection method for Quahog Parasite Unknown (QPX). Dis. Aquat. Org. 70:115-122. [DOI] [PubMed] [Google Scholar]
- 11.Gast, R. J., D. M. Moran, C. Audemard, M. M. Lyons, J. DeFavari, K. S. Reece, D. Leavitt, and R. Smolowitz. 2008. Environmental distribution and persistence of Quahog Parasite Unknown (QPX). Dis. Aquat. Org. 81:219-229. [DOI] [PubMed] [Google Scholar]
- 12.Gu, W., and R. E. Levin. 2006. Quantitative detection of Plesiomonas shigelloides in clam and oyster tissue by PCR. Int. J. Food Microbiol. 111:81-86. [DOI] [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Kleinschuster, S. J., R. Smolowitz, and J. Parent. 1998. In vitro life cycle and propagation of Quahog Parasite Unknown. J. Shellfish Res. 17:75-78. [Google Scholar]
- 15.Lyons, M. M., R. Smolowitz, C. F. Dungan, and S. B. Roberts. 2006. Development of a real time quantitative PCR assay for the hard clam pathogen Quahog Parasite Unknown (QPX). Dis. Aquat. Org. 72:45-52. [DOI] [PubMed] [Google Scholar]
- 16.Lyons, M. M., J. E. Ward, R. Smolowitz, K. R. Uhlinger, and R. J. Gast. 2005. Lethal marine snow: pathogen of bivalve mollusc concealed in marine aggregates. Limnol. Oceanogr. 50:1983-1988. [Google Scholar]
- 17.MacCallum, G. S., and S. E. McGladdery. 2000. Quahog Parasite Unknown (QPX) in the northern quahog Mercenaria mercenaria (Linnaeus, 1758) and M. mercenaria var. notata from Atlantic Canada, survey results from three maritime provinces. J. Shellfish Res. 19:43-50. [Google Scholar]
- 18.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 19.Prokopowich, C. D., T. R. Gregory, and T. J. Crease. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48-50. [DOI] [PubMed] [Google Scholar]
- 20.Qian, H., Q. Q. Liu, B. Allam, and J. L. Collier. 2007. Molecular genetic variation within and among isolates of QPX (Thraustochytridae), a parasite of the hard clam Mercenaria mercenaria. Dis. Aquat. Org. 77:159-168. [DOI] [PubMed] [Google Scholar]
- 21.Raghukumar, S. 2002. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistol. 38:127-145. [Google Scholar]
- 22.Ragone Calvo, L. M., J. G. Walker, and E. M. Burreson. 1998. Prevalence and distribution of QPX, Quahog Parasite Unknown, in hard clams, Mercenaria mercenaria in Virginia, USA. Dis. Aquat. Org. 33:209-219. [DOI] [PubMed] [Google Scholar]
- 23.Selesi, D., I. Pattis, M. Schmid, E. Kandeler, and A. Hartmann. 2007. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J. Microbiol. Methods 69:497-503. [DOI] [PubMed] [Google Scholar]
- 24.Skovhus, T. L., N. B. Ramsing, C. Holmstrom, S. Kjelleberg, and I. Dahllof. 2004. Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Appl. Environ. Microbiol. 70:2373-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smolowitz, R., D. Leavitt, and F. Perkins. 1998. Observations of a protistan disease similar to QPX in Mercenaria mercenaria (hard clams) from the coast of Massachusetts. J. Invertebr. Pathol. 71:9-25. [DOI] [PubMed] [Google Scholar]
- 26.Stokes, N. A., L. M. Ragone Calvo, K. S. Reece, and E. M. Burreson. 2002. Molecular diagnostics, field validation, and phylogenetic analysis of Quahog Parasite Unknown (QPX), a pathogen of the hard clam Mercenaria mercenaria. Dis. Aquat. Org. 52:233-247. [DOI] [PubMed] [Google Scholar]
- 27.Van der Auwera, G., S. Chapelle, and R. De Wachter. 1994. Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and phylogeny of the oomycetes. FEBS Lett. 338:133-136. [DOI] [PubMed] [Google Scholar]
- 28.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 29.Whyte, S. K., R. J. Cawthorn, and S. E. McGladdery. 1994. QPX (Quahaug Parasite X), a pathogen of northern quahaug Mercenaria mercenaria from the Gulf of St. Lawrence, Canada. Dis. Aquat. Org. 19:129-136. [Google Scholar]
- 30.Zhang, H., and S. J. Lin. 2005. Development of a cob-18S rRNA gene real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl. Environ. Microbiol. 71:7053-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, F., R. Massana, F. Not, D. Marie, and D. Vaulot. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79-92. [DOI] [PubMed] [Google Scholar]