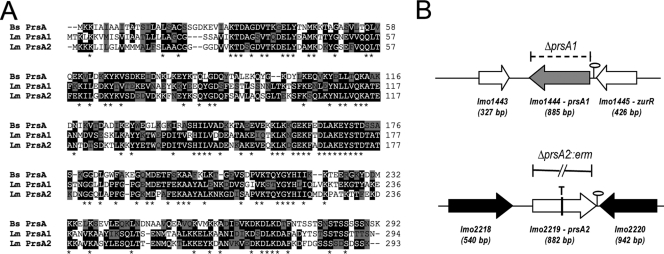
FIG. 1.
Amino acid sequence alignment and construction of prsA1 and prsA2 loss-of-function mutants. (A) Amino acid sequence alignment of B. subtilis PrsA and the two PrsA homologues in L. monocytogenes (PrsA1 and PrsA2) using the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw/index.html). Amino acid residues that are identical between at least two of the three PrsA proteins are shaded in black; asterisks indicate amino acid residues that are identical in all three proteins. Residues that are similar between protein sequences are shaded in gray. (B) Schematic of the ΔprsA1 deletion mutant, the prsA2::T targetron insertion mutant, and the ΔprsA2::erm mutant. The gray and white center arrows denote the open reading frames of prsA1 and prsA2, respectively, while the white and black flanking arrows denote flanking gene sequences. Predicted transcriptional terminators are depicted by stem-loops. The dashed bracket indicates the deletion of the prsA1 open reading frame, the vertical line through the lmo2219 open reading frame and T denote the intron insertion at nucleotide 457 in prsA2, and the solid broken line designates the ΔprsA2::erm deletion.