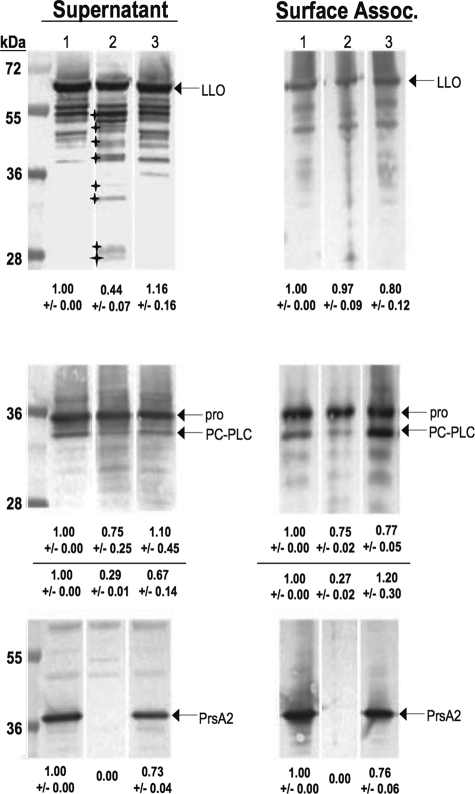
FIG. 6.
Strains lacking PrsA2 exhibit increased LLO degradation and reduced processing of pro-PC-PLC in the presence of activated PrfA. Bacterial supernatant and surface-associated proteins from the mutationally activated prfA(L140F), prfA(L140F) ΔprsA2::erm, or prfA(L140F) ΔprsA2::erm + pPL2-prsA2 mutant were isolated by TCA precipitation (secreted proteins) or boiling in 2% SDS (surface-associated proteins). Sample volumes were adjusted to reflect equivalent bacterial densities (OD600 of ∼1.4). Western analysis of protein fractions subjected to SDS-PAGE was done using rabbit polyclonal anti-LLO antibody, rabbit polyclonal anti-PrsA2, and rabbit polyclonal anti-PC-PLC antibody to detect LLO, PrsA2, and PC-PLC respectively. Arrows indicate bands of interest (either LLO, PrsA2, pro-PC-PLC, or PC-PLC) in each panel. Small stars indicate the altered LLO degradation pattern found in prfA(L140F) ΔprsA2::erm strains compared to that of prfA(L140F) and complement strains. The amount of protein detected for each sample in comparison to the WT lane (set at 1.0) as determined by densitometry is indicated at the bottom of each panel. The top and bottom rows of numbers located at the bottom of the PC-PLC Western blot represent the total amounts of pro-PC-PLC and PC-PLC, respectively. Lane 1, prfA(L140F); lane 2, prfA(L140F) ΔprsA2::erm; lane 3, prfA(L140F) ΔprsA2::erm + pPL2-prsA2.