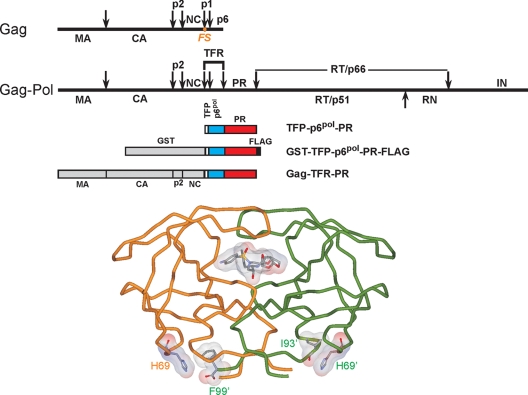
FIG. 1.
Structural organization of Gag and Gag-Pol polyproteins in HIV-1 and tube representation of the three-dimensional structure of the mature protease dimer in complex with the inhibitor DRV (Protein Data Bank accession number 2IEN [20]). Straight arrows indicate the specific sites of cleavage by the viral protease. The TFR, N-terminal to the protease, consists of the transframe octapeptide TFP and 48 amino acids of p6pol. The nomenclature of HIV-1 proteins is according to Leis at al. (9), as follows: CA, capsid; NC, nucleocapsid; RT, reverse transcriptase; RN, RNase H; and IN, integrase. Residues H69, F99, and I93 (proximal to H69) and DRV are shown as stick and surface representations. The protease precursor constructs used for monitoring the autocatalytic maturation reaction in vitro (TFR-PR) and in E. coli (GST-TFR-PR-FLAG) as well as the proviral construct expressed in 293T cells in vivo (Gag-TFR-PR) are all drawn to scale. The position of the frameshift (FS) that produces Gag-Pol is indicated as an orange dot. Calculated molecular weights of the domains indicated for the precursors are 26,583, 1,010, 5,357, 10,727, and 1,112 for GST, TFP, p6pol, PR, and FLAG, respectively. The proviral DNA construct contains the region for the expression of Gag and Gag-TFR-PR.