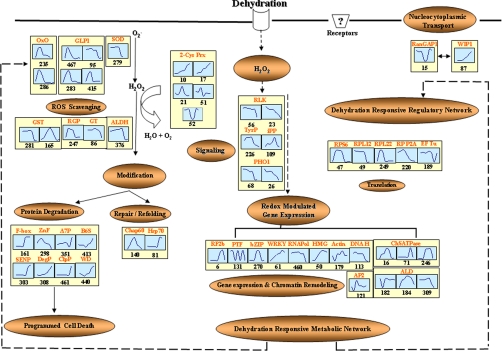
Fig. 11.
Pathway involved in dehydration-dependent functional and regulatory networks in rice nucleus. Proteins identified in this study are indicated in the box. Graphs are representative of expression profiles of individual proteins, and the number given below each graph indicates the protein identification number. OxO, oxalate oxidase; GLP1, germin-like protein 1; SOD, superoxide dismutase; ALDH, aldehyde dehydrogenase; 2-Cys Prx, 2-Cys peroxiredoxin; RLK, receptor-like protein kinase; TyrP, tyrosine phosphatase; iPP, inorganic pyrophosphatase; PHO1, PHO1-like protein; RPS6, ribosomal protein S6; RPL12, ribosomal protein L12; RPL22, ribosomal protein L22; RPP2A, ribosomal protein P2A; Chap60, chaperonin 60; Hsp70, 70-kDa heat shock protein; RGP, reversibly glycosylated peptide; GT, glycosyltransferase; F-box, F-box-like protein; ZnF, zinc finger, C3HC4 type (RING finger); A7P, α7 subunit of 20 S proteasome; B6S, β6 subunit of 20 S proteasome; DegP, putative DegP protease; SENP, putative sentrin-specific protease; ClpP, ATP-dependent Clp protease proteolytic subunit; WD, WD repeat-containing protein, putative; RF2b, bZIP transcription factor RF2b; PTF, putative transcription factor; bZIP, putative bZIP protein; WRKY, WRKY transcription factor; RNApol, putative RNA polymerase II subunit 5; HMG, putative high mobility group I/Y; Actin, putative actin; ALD, fructose-bisphosphate aldolase; DNA H, putative DNA helicase; ChSATPase, chromosome segregation ATPases; AP2, AP2 domain-containing protein; WIP, WPP domain-interacting protein 1; RanGAP1, Ran GTPase-activating protein 1.