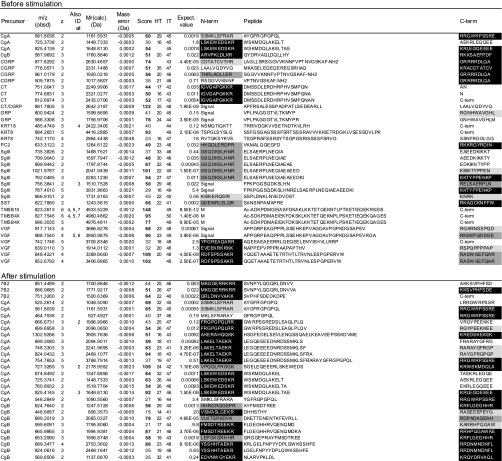
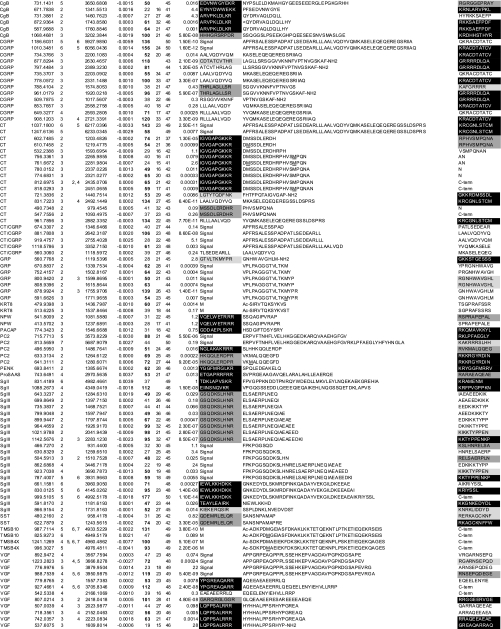
Table I. Peptides identified before and after stimulation.
Data were obtained without gel filtration chromatography and summarized from three runs using an identical LC-MSMS setting and peptides whose scores (column 7) exceeded homology thresholds (HT, column 8) in at least two runs are listed. For peptides identified in multiple runs, a higher score is listed. Mr(Calc) represents the theoretical monoisotopic molecular mass (Da) based on the peptide sequence. The score value beyond an identity threshold (IT, column 9) is indicated in bold. In column 1, “CT/CGRP” indicates that the peptides are shared by CT and CGRP precursors. If the peptide was identified at different charges, the charge states are also shown in column 4. Expectation values are indicated in column 10. The N- (N-term) and C-terminal (C-term) flanking 10 amino acids (columns 11 and 13) are shown and marked as follows: closed boxes with white letters, typical PC cleavage sites containing consecutive basic residues and sites having C-terminal amidation motifs; dark gray boxes, cleavage sites containing basic residues at P4, P6 or P8 position; pale gray boxes, sites having basic residues at P1 but not at P2, P4, P6 or P8 position. “Signal” indicates that the peptide flanks its signal sequence. “C-term” indicates that the peptide C-terminus is the end of the precursor protein. Ac-, N-terminal acetylation; -NH2, C-terminal amidation; <Q, pyroglutamic acid. Oxidized methionine residues are underlined in column 12. Sequences are based on the following IPI accession numbers: CgA, 00746813; CgB, 00006601, CGRP, 00027855; CT, 00000914; GRP, 00011722; KRT18, 00554788; KRT8, 00554648; PC2, 00029131; SgIII, 00292071; SST, 00000130; TMSB10, 00220827; TMSB4X, 00220828; VGF, 00069058; 7B2, 00008944; NPW, 00853190; PACAP, 00000027; PENK, 00000828; SgII, 00009362. PACAP, pituitary adenylate cyclase-activating polypeptide; NPW, neuropeptide W; PENK, proenkephalin A; KRT, cytokeratin; TMSB4X, thymosin β-4 X-linked; TMSB10, thymosin β-10.