Abstract
Diagnosis of lysosomal storage diseases (LSDs) can be problematic in atypical cases where clinical phenotype may overlap with other genetically distinct disorders. In addition, LSDs may result from mutations in genes not yet implicated in disease. Thus, there are individuals that are diagnosed with apparent LSD based upon clinical criteria where the gene defect remains elusive. The objective of this study was to determine whether comparative proteomics approaches could provide useful insights into such cases. Most LSDs arise from mutations in genes encoding lysosomal proteins that contain mannose 6-phosphate, a carbohydrate modification that acts as a signal for intracellular targeting to the lysosome. We purified mannose 6-phosphorylated proteins by affinity chromatography and estimated relative abundance of individual proteins in the mixture by spectral counting of peptides detected by tandem mass spectrometry. Our rationale was that proteins that are decreased or absent in patients compared with controls could represent candidates for the primary defect, directing biochemical or genetics studies. On a survey of brain autopsy specimens from 23 patients with either confirmed or possible lysosomal disease, this approach identified or validated the genetic basis for disease in eight cases. These results indicate that this protein expression approach is useful for identifying defects in cases of undiagnosed lysosomal disease, and we demonstrated that it can be used with more accessible patient samples, e.g. cultured cells. Furthermore this approach was instrumental in the identification or validation of mutations in two lysosomal proteins, CLN5 and sulfamidase, in the adult form of neuronal ceroid lipofuscinosis.
The genetic bases for numerous human hereditary diseases are well established (1), but there are some for which the defective genes remain to be identified. For example, in the Online Mendelian Inheritance in Man database (2), there are currently listed over a thousand Mendelian clinical phenotypes of unknown molecular basis. Understanding the molecular basis for disease is essential for genetic screening and developing effective therapy, but identifying individual gene defects can represent a significant challenge. This is particularly true of orphan diseases where patient populations may be small or clinically poorly defined and thus may not be readily amenable to traditional genetics approaches. Proteomics methods provide an alternative route in the investigation of such unsolved genetic diseases and can provide disease gene candidates for further analysis in two different ways. First, comparative proteomics can uncover proteins that are altered in abundance or other properties in specimens from affected individuals, and these may potentially be encoded by the mutant gene. Second, descriptive proteomics can identify novel proteins with known or predicted properties or expression patterns that may associate them with diseases of unknown etiology. Such approaches can be applicable to small cohorts or even individual cases and have been particularly useful in the investigation of lysosomal storage diseases (LSDs)1 (for a review, see Ref. 3).
LSDs most commonly result from mutations in genes encoding lysosomal enzymes and accessory proteins. As one of the primary functions of the lysosome is the degradation of cellular macromolecules (for a review, see Ref. 4), a hallmark of these diseases is an accumulation of undigested substrates and other storage material within the lysosomes in cells of affected individuals. Most LSD genes have been identified using classical biochemical or genetics methods, but there are some clinically defined LSDs for which the defective genes are not known. Proteomics approaches have been instrumental in the discovery of gene defects in three such human diseases. For Niemann-Pick C type 2 disease (5) and mucopolysaccharidosis (MPS) IIIC (6), pathogenic mutations were found in genes encoding novel lysosomal proteins that were discovered in proteomics surveys. In classical late infantile neuronal ceroid lipofuscinosis (LINCL), the mutant gene product was identified as a spot that was present on two-dimensional gels of extracts from brain autopsy specimens of controls but not of affected individuals (7).
In addition to these clinically defined but unsolved diseases, there are individual cases with histopathological evidence of lysosomal storage where gene defects have not been identified. There are several possible reasons for the lack of a definitive genetic diagnosis in these cases. First, given the heterogeneity and overlap in clinical presentation of LSDs, especially when dealing with atypical alleles, no obvious underlying gene defect may be apparent. Second, genetic validation may be difficult to establish if mutations occur outside of coding regions or if they cannot be distinguished from harmless polymorphisms. Third, disease may result from mutations in genes that encode proteins that are not currently thought to be associated with LSDs.
In this study, we conducted a comparative analysis of purified mannose 6-phosphate (Man-6-P) glycoproteins from brain autopsy samples from potential LSD cases of unsolved or ambiguous etiology to investigate the molecular basis of disease. Our rationale was that we may identify lysosomal proteins that are altered in terms of expression in cases compared with controls and that this information may provide valuable clues to the cause of disease. Central to this approach is the ability to selectively isolate lysosomal proteins for analysis. Most newly synthesized lysosomal enzymes are glycoproteins that receive a specialized carbohydrate modification, Man-6-P, that is recognized by Man-6-P receptors (MPRs). The MPRs bind both newly synthesized lysosomal proteins in the Golgi and extracellular lysosomal proteins at the cell surface, and they travel to an acidic endolysosomal compartment where the low pH directs dissociation of receptor and ligand. From an analytical perspective, immobilized MPRs can be used to affinity purify proteins containing Man-6-P, and this has facilitated numerous studies of the lysosomal proteome (for a review, see Ref. 8).
In this report, we demonstrate that mass spectrometry-based proteomics profiling can successfully lead to a definitive genetic diagnosis in difficult cases where conventional methods were not fruitful. In addition to providing a proof of principle for a clinical proteomics approach to LSDs, this approach was instrumental in the discovery of two lysosomal proteins in which defects may underlie the adult form of NCL, a neurodegenerative disease of currently uncertain basis.
EXPERIMENTAL PROCEDURES
Human Brain Autopsy Samples
Research protocols involving human subjects were approved by the Institutional Review Board of the University of Medicine and Dentistry of New Jersey. Patient information is summarized in Table I. Brain autopsy samples with HSB identifiers were obtained from the Human Brain and Spinal Fluid Resource Center (Los Angeles, CA), samples with UMB identifiers were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD), and samples with CABM identifiers were generously referred to us by other researchers. Typically cerebrum sections frozen in dry ice/alcohol baths were used for this study, and where known, the average post-mortem interval was 10.5 h with a range of 4–22 h. Brain samples were stored at −80 °C until use.
Table I. Summary of patient samples.
Clinical data are provided where available. M, male; F, female, n/a, not applicable.
| Identifier | Diagnosis | Age of onset | Age of death | Post-mortem interval | Gender |
|---|---|---|---|---|---|
| years | years | h | |||
| HSB 120 | Adenoleukodystrophy | 56 | M | ||
| CABM-BR19 | Adult NCL | 20 | 33 | F | |
| CABM-BR8 | Adult NCL | Late 30s | 50s | M | |
| HSB 2356 | Adult NCL | 33 | 40 | 8 | M |
| HSB 4165 | Adult NCL | 29 | 53 | 6.5 | M |
| UMB 338 | Alexander disease | 6.2 | 12 | M | |
| UMB 613 | Alexander disease | 14 | 7 | M | |
| UMB 575 | Krabbe disease | 1.4 | 4 | M | |
| CABM-BR21 | Late infantile NCL (CLN2) | M | |||
| UMB 784 | Late infantile NCL (CLN2) | 5.3 | 4 | F | |
| UMB 563 | Mucopolysaccharidosis IIIA | 11.3 | 12 | F | |
| UMB 821 | Neuromuscular disorder, undiagnosed | 3.6 | 8 | F | |
| HSB 238 | Normal control | n/a | 43 | F | |
| HSB 298 | Normal control | n/a | 19 | M | |
| HSB 304 | Normal control | n/a | 22 | M | |
| HSB 331 | Normal control | n/a | 16 | M | |
| HSB 712 | Normal control | n/a | 16 | M | |
| HSB 863 | Normal control | n/a | 40 | M | |
| UMB 1270 | Seizure disorder | 22 | 22 | F | |
| UMB 506 | Sudanophilic leukodystrophy | 0.4 | 5 | F | |
| HSB 148 | Tay-Sachs disease | 0.6 | 1.9 | F | |
| HSB 1183 | Unknown epilepsy | 0.2 | 1 | F | |
| CABM-BR1 | Unknown NCL | M | |||
| CABM-BR11 | Unknown NCL | F | |||
| CABM-BR23 | Unknown NCL | ||||
| CABM-BR24 | Unknown NCL | 24 | 20.6 | F | |
| CABM-BR25 | Unknown NCL | 8 | 16.5 | F | |
| CABM-BR32 | Unknown NCL | F | |||
| CABM-BR33 | Unknown NCL | 12.4 | F | ||
| CABM-BR34 | Unknown NCL | 7.5 | M | ||
| CABM-BR36 | Unknown NCL | F | |||
| HSB 4391 | Unknown NCL | 12 | F | ||
| HSB 2548 | Variant late infantile NCL | 7 | 25 | F |
Purification of Man-6-P Glycoproteins
Reagents were from Sigma unless noted otherwise. Man-6-P glycoproteins were purified from brain essentially as described previously (9). In brief, 1–2 g of brain was thawed and homogenized using a Polytron homogenizer (Brinkmann Instruments) in 100 ml of homogenization buffer that comprised PBS with inhibitors (I) to prevent proteolysis and dephosphorylation (2.5 mm EDTA, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 0.25 mm Pefabloc (Pentafarm, Basel, Switzerland), 5 mm β-glycerophosphate) and 0.2% Tween 20 (T). The homogenate was centrifuged at 25,000 × g for 1 h at 4 °C, and the supernatant was filtered through Whatman 3M filter paper. The filter paper was washed with an additional 30 ml of homogenization buffer, and the pooled filtrate was loaded overnight onto 4-ml-bed volume affinity columns of soluble cation-independent MPR immobilized at a density of 2.5 mg/ml Affi-Gel 15 (Bio-Rad). Columns were flow-washed with 30 ml of PBS-TI and then batch-washed three times with 10 ml of PBS-TI followed by three times with the same homogenization buffer without detergent (PBS-I). Columns were flow-washed overnight with an additional 80 ml (20 bed volumes) of PBS-I. Columns were sequentially batch-eluted with 8 ml of each of the following: PBS; PBS containing 10 mm mannose, 10 mm glucose 6-phosphate (mock eluate); and PBS containing 10 mm mannose 6-phosphate (specific eluate). Eluates were concentrated to a final volume of ∼100 μl using Centricon 10 spin concentrators (Millipore, Billerica, MA), and protein concentration was determined using dye-binding reagent (10). Affinity columns were reequilibrated with PBS and stored in PBS containing 0.05% sodium azide prior to reuse.
Purification of Man-6-P Glycoproteins from Cultured Human Cells
Immortalized lymphoblasts and primary fibroblasts were grown for 6 days in RPMI 1640 culture medium (Invitrogen) containing fetal bovine sera (5 and 10%, respectively), penicillin-streptomycin, and 30 μm chloroquine (Sigma), a lysosomotropic agent that induces the secretion of lysosomal proteins that retain Man-6-P (11). Conditioned media were harvested by centrifugation and supplemented with phosphatase and protease inhibitors, and affinity purification of Man-6-P glycoproteins on immobilized soluble cation-independent MPR was performed as described above.
Tandem Mass Spectrometry
Five micrograms of each sample was run ∼1 cm by SDS-PAGE and excised as a single gel slice to facilitate subsequent handling and in-gel tryptic digestion (9). LC-MS/MS experiments were performed using a Dionex U3000 nanoflow chromatography system in line with an LTQ linear ion trap mass spectrometer (ThermoFisher, Waltham, MA). Duplicate samples (1-μg equivalents) of each digest were analyzed using LC-MS/MS with a 3-h gradient as described previously (12). Briefly each full-scale MS scan was followed by 10 MS/MS scans on the most abundant precursor ions with precursor ions that were fragmented twice within a 30-s interval being excluded from further analysis for a period of 60 s. Additional experiments to confirm the deficiency of select proteins using targeted MS/MS were conducted in a similar manner except that six precursor ions, three from the protein of interest and three from a control, were chosen for MS/MS. In these experiments, 1 μg of each digest was again analyzed as described previously (12), but the acquisition method was set to perform one MS scan from 300 to 2000 m/z followed by MS/MS of the six select masses. Peptide abundance was estimated by determining the peak areas of three prominent fragment ions derived from each selected peptide.
Database Searching
Peak lists were generated from raw LTQ data using the TurboSEQUEST module of Bioworks 3.3 (Thermo Electron). Parameters were essentially as described previously (12) except that a charge state of +2 was specified when generating the dta files from each LC-MS/MS run that were then merged into an mgf file. Subsequent database searching was then conducted allowing matches against charge states +1, +2, and +3 for each precursor. This procedure reduces the size of the mgf files compared with those created using automatic charge state determination (+2 and +3 charge states assigned to each ion) and eliminates the rare cases where a single MS/MS event results in generation of two dta files that are matched to two different peptides. Tandem mass spectrometry data were searched against the 49.36 version of the NCBI 36 assembly of the human genome using a local implementation of Global Proteome Machine Extreme Edition Manager version 2.2.1 (Beavis Informatics Ltd., Winnipeg, Canada) that uses the Tornado (2008.01.01) version of X!Tandem to assign spectral data (38). Searches were conducted using a precursor ion mass error of +4 and −0.5 Da and a fragment mass error of 0.4 Da. Errors in assignment of monoisotopic mass were not permitted. Trypsin cleavage was defined to be C-terminal to every lysine or arginine except when followed by a proline, and up to one missed cleavage site was allowed. Cysteine carbamidomethylation was specified as a fixed modification, and methionine oxidation was a permitted variable modification during development of the preliminary model. Methionine oxidation and deamidation at asparagine and glutamine residues were allowed during model refinement of those preliminary assignments that achieved an expectation value of <10−6. Database searching was conducted using a multidimensional protein identification technology approach with the GPM as described previously (9) where mgf files from all LC-MS/MS runs were used in the search to generate a single concatenated output from which individual sample data could subsequently be extracted. This approach allows for a consistent assignment between different LC-MS/MS runs of those spectra that can be assigned to multiple gene products. To avoid situations where peptides may be assigned to multiple protein isoforms derived from the same gene (e.g. splice variants), protein identifiers in the GPM output (Ensembl ENSP) were converted to gene identifiers (Ensembl ENSG) prior to analysis. Criteria for protein identification were at least two unique peptide assignments and a minimum log expectation score of −10. (The log(e) score is the base 10 log expectation score that the protein or peptide assignment is stochastic.) Mass spectrometry data were quantified by spectral counting (13, 14).
Statistical Analysis of Spectral Count Data
Relative protein abundance in mock and specific affinity column eluates was used to differentiate between proteins that were specifically purified (i.e. enriched in the specific eluate) and nonspecific contaminants that are equally abundant in both (9). The method of Wilson (39) was used to calculate the upper and lower limits of the 95% confidence interval for the ratio of spectral counts found in the specific compared with mock eluate. Analyses were conducted using R version 2.5.0.
DNA Sequencing
Genomic DNA was prepared using the Qiagen (Valencia, CA) Blood and Tissue kit, and exonic PCR products were sequenced using ABI BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) on an ABI 3130xl Genetic Analyzer at the Molecular Resource Facility of the New Jersey Medical School.
RESULTS
Brain Autopsy Samples
Thirty-three human brain autopsy samples were obtained, representing normal controls (n = 6), negative control cases with known defects that do not involve lysosomal Man-6-P glycoproteins (Alexander disease; n = 2), positive control cases with defects in lysosomal proteins that have been determined by biochemical and/or genetics methods (MPSIIIA, Tay-Sachs disease, classical LINCL, and Krabbe disease; n = 5), and cases with possible lysosomal disease of unknown or incompletely defined basis (n = 20). Case information is summarized in Table I.
Strategy for the Identification of Gene Defects
The rationale underlying this study was that a mass spectrometric proteomics analysis of lysosomal proteins could reveal those that are absent, diminished, or otherwise altered in LSD cases compared with controls. As candidates for the product of the mutant gene, such proteins could be investigated further using biochemical and genetics methods. Soluble lysosomal proteins containing Man-6-P were purified by MPR affinity chromatography prior to analysis (15).
This approach required the ability to compare relative protein levels in control and case samples. This was achieved by analysis of samples by data-dependent LC-MS/MS. These data were used to construct a spectral count index (13, 14) (the number of MS/MS spectra that could be assigned to a given protein in a given sample) to estimate the relative abundance of proteins in control and disease specimens. We then performed targeted MS/MS on select peptides from control and candidate proteins to verify that the latter are actually depleted or absent and not missed for technical reasons. (For example, ion suppression from peptides derived from a protein that is elevated in a given sample could result in decreased spectral counts for another protein if one or more peptides from the two proteins co-elute during LC.) Finally genetics methods and, in some cases, biochemical assays were used to investigate potential candidates.
Protein assignments, including expectation probability scores and protein coverage, are shown in supplemental Table 1. When data for all samples are considered together, 320 proteins met our criteria for assignment. Of these, 58 were known lysosomal proteins that are known to contain Man-6-P.
An inexact diagnosis could reflect atypical clinical presentation due to variant or late onset alleles in known lysosomal proteins, and we expected our approach to readily identify such deficiencies. However, we also considered the possibility that disease may result from defects in Man-6-P glycoproteins that are not currently assigned to the lysosome but that actually do reside within this organelle. Numerous proteins that are not currently assigned to the lysosome were identified in our preparations from human brain, and some of these represent bona fide Man-6-P glycoproteins that reside within this organelle. It is important to consider such proteins in our analysis, but they must be distinguished from contaminants, e.g. abundant or sticky proteins that are not completely removed by the affinity purification. This can be achieved by comparing the relative abundance of proteins that are eluted from the MPR affinity column with either a mixture of glucose 6-phosphate and mannose (mock eluate) or Man-6-P (specific eluate) with the rationale that proteins containing Man-6-P will be enriched in the latter compared with the former. We have previously used this approach to identify Man-6-P glycoproteins purified from rat tissues (9), and here we considered the human orthologs of such rat proteins in our analysis. We also performed a similar study on the preparations from control human brains to directly identify such proteins (supplemental Table 2). Of the 58 identified known lysosomal proteins, 57 were found to be significantly enriched in the specific eluate. The remaining lysosomal protein was present only in the specific eluate but at levels that were too low to allow confident conclusions regarding distribution. Of the 282 identified proteins that are not currently assigned to the lysosome, 31 were enriched in the specific eluate and are therefore of interest as potential Man-6-P glycoproteins. Of the remainder, no confident conclusion could be made for 90 proteins, whereas 161 were significantly enriched in the mock eluate, indicating that they are likely to be contaminants.
Expression of Lysosomal Proteins in Human Brain
The confidence that any given protein may be deficient in a patient sample is related to the abundance of this protein in control samples because the probability that a protein may be missed through sampling variation or natural heterogeneity increases as abundance decreases. (The example of galactosylceramidase discussed below clearly illustrates this problem.) We therefore set an arbitrary minimum threshold of an average of 10 spectral counts to identify proteins whose absence would provide a convincing argument for further investigation. Representative spectral count data are shown for several lysosomal proteins for each sample in Table II, and the complete data set for both lysosomal and non-lysosomal proteins is provided in supplemental Table 3.
Table II. Representative spectral count data for select lysosomal proteins.
Bold values indicate proteins encoded by mutant genes in the respective case. Counts are normalized by multiplying by the average number of spectra assigned in all samples divided by the total number of spectra assigned in that particular sample. Bold indicates verified defects. HEXA, β-hexosaminidase α.
| Identifier | Select lysosomal proteins | ||||||
|---|---|---|---|---|---|---|---|
| CTSD | ASAH1 | PPT1 | HEXA | TPP1 | SGSH | CLN5 | |
| HSB 238 | 1054.0 | 231.0 | 246.8 | 83.3 | 51.6 | 9.5 | 12.7 |
| HSB 298 | 898.6 | 174.3 | 143.5 | 117.0 | 25.6 | 29.9 | 12.8 |
| HSB 304 | 1207.5 | 191.9 | 263.2 | 85.7 | 44.0 | 15.9 | 6.8 |
| HSB 331 | 1190.2 | 191.4 | 211.9 | 86.3 | 39.3 | 14.4 | 8.3 |
| HSB 712 | 1049.6 | 196.6 | 170.8 | 76.3 | 36.3 | 21.0 | 13.4 |
| HSB 863 | 765.9 | 312.5 | 229.3 | 82.2 | 79.2 | 23.6 | 7.2 |
| CABM-BR1 | 903.8 | 440.9 | 169.3 | 103.3 | 74.6 | 48.8 | 14.3 |
| CABM-BR11 | 537.3 | 350.6 | 399.7 | 93.4 | 109.8 | 80.3 | 9.0 |
| CABM-BR19 | 969.5 | 271.0 | 228.9 | 98.8 | 41.1 | 30.3 | 0.0 |
| CABM-BR21 | 303.3 | 563.7 | 145.7 | 102.7 | 1.2 | 60.9 | 21.5 |
| CABM-BR23 | 744.4 | 160.2 | 200.6 | 140.0 | 127.9 | 59.2 | 5.4 |
| CABM-BR24 | 348.9 | 157.6 | 150.4 | 185.3 | 30.1 | 99.9 | 26.5 |
| CABM-BR25 | 418.9 | 127.2 | 148.6 | 107.0 | 0.0 | 82.2 | 16.9 |
| CABM-BR32 | 734.2 | 441.1 | 391.1 | 82.6 | 286.4 | 44.2 | 5.8 |
| CABM-BR33 | 778.0 | 322.3 | 309.3 | 107.4 | 54.3 | 41.3 | 20.1 |
| CABM-BR34 | 636.1 | 473.1 | 203.0 | 53.0 | 90.0 | 22.0 | 42.0 |
| CABM-BR36 | 617.3 | 376.1 | 148.8 | 104.4 | 133.1 | 13.9 | 11.1 |
| CABM-BR8 | 922.3 | 300.4 | 398.7 | 74.4 | 60.6 | 33.1 | 21.1 |
| HSB 1183 | 673.5 | 480.8 | 220.0 | 114.5 | 140.6 | 11.3 | 18.1 |
| HSB 120 | 719.5 | 485.4 | 286.5 | 81.3 | 249.1 | 24.6 | 7.5 |
| HSB 148 | 1058.7 | 134.8 | 291.4 | 0.0 | 203.6 | 8.7 | 9.6 |
| HSB 2356 | 771.2 | 280.4 | 419.0 | 90.7 | 67.7 | 33.4 | 15.6 |
| HSB 2548 | 527.9 | 322.9 | 325.4 | 82.0 | 235.9 | 38.5 | 20.1 |
| HSB 4165 | 898.2 | 238.0 | 369.3 | 87.1 | 87.1 | 0.0 | 20.9 |
| HSB 4391 | 1388.2 | 306.7 | 105.6 | 102.9 | 0.9 | 32.4 | 24.1 |
| UMB 1270 | 333.2 | 268.6 | 212.9 | 124.3 | 92.5 | 35.8 | 22.9 |
| UMB 338 | 872.2 | 385.3 | 265.0 | 131.4 | 86.1 | 16.6 | 16.6 |
| UMB 506 | 840.7 | 441.4 | 147.5 | 110.4 | 43.1 | 18.1 | 19.1 |
| UMB 563 | 1074.0 | 262.5 | 545.9 | 107.1 | 117.6 | 0.0 | 17.8 |
| UMB 575 | 901.3 | 393.5 | 151.2 | 130.3 | 94.7 | 17.2 | 17.2 |
| UMB 613 | 203.0 | 118.2 | 56.6 | 168.1 | 10.0 | 84.9 | 15.0 |
| UMB 784 | 702.6 | 191.7 | 265.8 | 141.6 | 10.9 | 40.3 | 17.4 |
| UMB 821 | 872.9 | 405.1 | 217.3 | 130.9 | 86.3 | 19.9 | 23.7 |
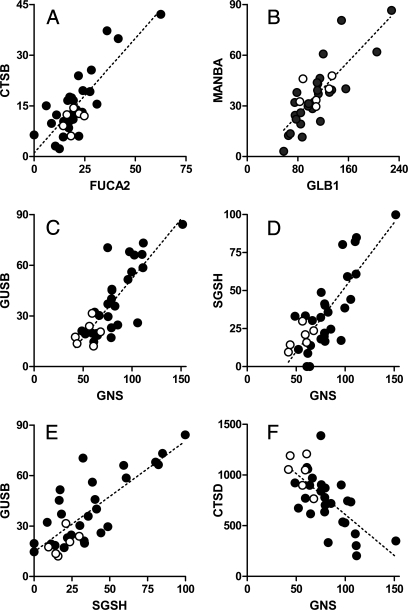
Lysosomal hydrolases frequently catalyze sequential steps in the degradation of macromolecules, and some enzymes assemble into multiprotein complexes; thus it is possible that expression of some of these proteins may be co-regulated. To investigate this possibility, we performed a correlation analysis of relative abundance in brain (supplemental Table 4), and we found that the relative amounts of some lysosomal proteins are significantly related (Fig. 1). Interestingly proteins that showed the highest correlation tend to function in related catabolic pathways. Chitobiase and plasma α-fucosidase (Fig. 1, Panel A) as well as β-galactosidase and β-mannosidase (Fig. 1, Panel B) are involved in the degradation of glycoproteins. Expression levels of β-glucuronidase, glucosamine (N-acetyl)-6-sulfatase, and N-sulfoglucosamine sulfohydrolase (SGSH; sulfamidase) are interrelated (Fig. 1, Panels C–E), and all three play roles in the degradation of glycosaminoglycans. These data suggest that expression of some lysosomal proteins is coordinately regulated and that this approach may potentially provide useful insights into their functional interactions. In some cases, correlation of expression may reflect protein-protein interactions that occur during purification. For example, expression of the relatively abundant lysosomal cysteine protease cathepsin Z was correlated with that of a non-lysosomal cysteine protease inhibitor, cystatin B, which does not contain Man-6-P (data not shown). This correlation is not likely to be biologically meaningful as the two proteins probably associate after tissue disruption and thus co-purify. It is also possible that levels of a protein would be inversely correlated with that of another involved in its turnover, and this may be the case with glucosamine (N-acetyl)-6-sulfatase and the lysosomal protease cathepsin D (CTSD; Fig. 1, Panel F).
Fig. 1.
Coordinated expression of lysosomal proteins. Panels represent the following protein pairs: Panel A, chitobiase (CTBS) and plasma α-fucosidase (FUCA2), r2 = 0.673; Panel B, β-mannosidase (MANBA) and β-galactosidase (GLB1), r2 = 0.662; Panel C, β-glucuronidase (GUSB) and glucosamine (N-acetyl)-6-sulfatase (GNS), r2 = 0.655; Panel D, SGSH and glucosamine (N-acetyl)-6-sulfatase, r2 = 0.648; Panel E, β-glucuronidase and SGSH, r2 = 0.640; Panel F, CTSD and glucosamine (N-acetyl)-6-sulfatase, r2 = 0.480. For all plots, the linear relationship when considering controls and cases in aggregate was statistically significant (p < 0.0001), and the strength of the respective correlation is indicated by r2 values calculated from the Pearson correlation coefficient. Each point represents the spectral counts of the indicated two proteins associated with a given specimen with the raw data presented in supplemental Table 3. Filled and unfilled circles represent cases and controls, respectively.
Identification of Lysosomal Defects by Comparative Mass Spectrometry
We analyzed samples from positive control cases with biochemically and/or genetically confirmed LSD (Tay-Sachs disease, Krabbe disease, classical LINCL, and MPSIIIA) and negative controls with confirmed non-lysosomal defects (Alexander disease). In terms of our experimental cohort, we investigated cases with a clinically defined but genetically ambiguous diagnosis (e.g. unspecified NCL, adult NCL) and cases with an unclear diagnosis that could possibly be lysosomal in origin (e.g. undiagnosed LSD and others). Results are grouped according to diagnosis subsequent to our analysis.
Krabbe Disease
In the sample of Man-6-P glycoproteins prepared from brain from a Krabbe disease patient (UMB 575), we found that galactocerebrosidase could not be detected by LC-MS/MS, and this is consistent with diagnosis. However, we also failed to detect galactosylceramidase in other samples where other genetic defects had already been confirmed (supplemental Table 2). This reflects the low abundance of galactosylceramidase in brain and the correspondingly low numbers of spectral counts obtained for this protein.
Mucopolysaccharidosis IIIA
Case UMB 563 was originally obtained with a description of Sanfilippo syndrome, which encompasses four genetically distinct disorders. Spectral counting (Table II) and targeted MS (data not shown) of purified Man-6-P glycoproteins indicated a loss of peptides corresponding to SGSH in this case. Subsequent genetics analysis (Table III) then confirmed homozygosity for 746G→A (R245H), which is a common pathogenic allele in MPSIIIA (16). Subsequent clinical follow-up for this case revealed that this patient was in fact originally diagnosed with MPSIIIA (OMIM 252900); therefore this case was reclassified as a positive control with a known defect (Table III).
Table III. Summary of cases in which the comparative proteomics approach provided informative data.
Note that for each case the original diagnosis and its clinical basis are shown. Also indicated are biochemical and/or genetics analyses that validate the final diagnosis. GALC, galactosylceramidase; HEXA, β-hexosaminidase α.
| Identifier | Diagnosis (basis) | Informative lysosomal protein by comparative MS | Supporting data | Allele 1 | Allele 2 |
|---|---|---|---|---|---|
| Positive controls (known lysosomal defect) | |||||
| UMB 563 | MPSIIIA (unknown criteria, not genetic) | SGSH | Genetic | SGSH 746G→A (R245H) | SGSH 746G→A (R245H) |
| HSB 148 | Pediatric CNS/Tay-Sachs disease (enzyme assay) | HEXA | Enzyme assay | ||
| UMB 784 | Classical LINCL (ultrastructural pathology) | TPP1 | Enzyme assay, genetic | TPP1 3556G→C (splicing defect) | TPP1 3556G→C (splicing defect) |
| CABM-BR21 | Classical LINCL (enzyme assay and genetics analysis) | TPP1 | Enzyme assay, genetic | TPP1 3556G→C (splicing defect) | TPP1 3670C→T (R208Stop) |
| UMB 575 | Krabbe disease (unknown criteria) | GALC | |||
| Genetic basis undetermined | |||||
| CABM-BR25 | Undiagnosed NCL | TPP1 | Enzyme assay, genetic | TPP1 3016T→A (L104Stop) | TPP1 3556G→C (splicing defect) |
| HSB 4391 | Undiagnosed LSD | TPP1 | Enzyme assay, genetic | TPP1 deletion T2979 | TPP1 3084C→T (R127Stop) |
| CABM-BR19 | Adult NCL (ultrastructural pathology) | CLN5 | Genetic | CLN5 377G→A (C126Y) | CLN5 1121A→G (Y374C) |
| HSB 4165 | Adult NCL (ultrastructural pathology) | SGSH | Genetic | SGSH 904T→C (S298P) | SGSH 1075G→A (E355K) |
Classical Late Infantile Neuronal Ceroid Lipofuscinosis
We investigated two positive control cases that had previously been diagnosed with classical LINCL, which is due to mutations in the gene encoding tripeptidyl-peptidase 1 (TPP1). In both cases, diagnosis was confirmed by biochemical assay confirming a loss of TPP1 activity and by genetics analysis (Table III). In a patient originally diagnosed based upon ultrastructural pathology (UMB 784), peptides corresponding to TPP1 were present at reduced levels (∼10%) compared with other samples. In another case (CABM-BR21), we found that TPP1 was almost completely absent (∼1% of average levels in other samples). Two other cases (CABM-BR25 and HSB 4391) were obtained with an ambiguous diagnosis, i.e., unspecified NCL or Batten disease. In both, spectral counting indicated an apparent loss of TPP1, and this was confirmed by subsequent measurement of TPP1 activity and genetics analysis (Table III). These results indicate that these two cases actually represent classical LINCL.
Tay-Sachs Disease
We analyzed Man-6-P glycoproteins from a patient classified with pediatric CNS/Tay-Sachs disease (HSB 148) and found that the normally abundant lysosomal protein β-hexosaminidase α was absent. This finding supports the diagnosis of Tay-Sachs disease that was based on an absence of detectable serum β-hexosaminidase α activity (HSB 148 pathology report).
Adult Neuronal Ceroid Lipofuscinosis (ANCL)
ANCL, also known as Kuf disease (OMIM 204300 and OMIM 162350) is a rare neurodegenerative LSD of unknown genetic etiology. In this study, we investigated four cases diagnosed with this disease and identified genetic defects in two.
For patient CABM-BR19, a described ANCL case (17), a genetic screen was previously initiated focusing on known NCL genes to determine whether disease in this patient represented an atypical late onset variant form of another NCL.2 Two missense changes (377G→A (C126Y) and 1121A→G (Y374C)) were found in CLN5,2 a soluble lysosomal protein of unknown function that is defective in one of the variant forms of LINCL (18).
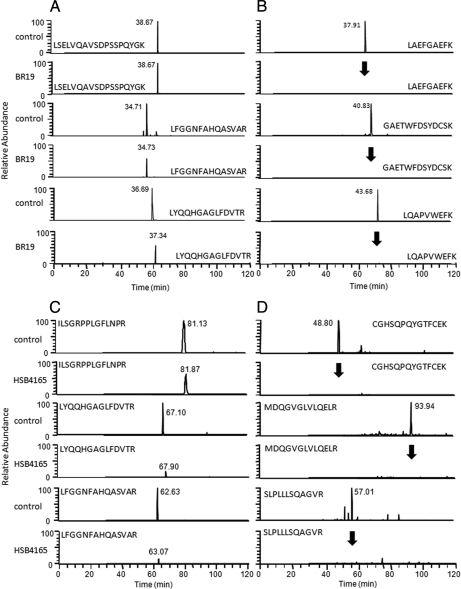
Here we reanalyzed this purified Man-6-P glycoprotein preparation using LC-MS/MS and spectral count analysis to determine whether alterations in CLN5 levels were associated with the missense changes. CLN5 was present in all of the normal control and disease cases but could not be detected in this ANCL sample (Table II). To determine whether CLN5 was actually absent in this case or whether there was an apparent loss that was technical in nature (e.g. due to peptide ion suppression or due to levels being below the detection threshold for data-dependent acquisition on the linear ion trap), we performed targeted mass spectrometry for select CLN5 peptides in control and CABM-BR19 samples (Fig. 2, Panels A and B). Although peptides corresponding to a control protein (TPP1) were present in both the control and case samples, the three peptides corresponding to CLN5 were absent in the latter. These results demonstrate that the missense changes identified by genetics analysis result in a loss of CLN5 and, given that a loss of this protein is typically associated with NCL disease, provide convincing evidence that they are pathogenic alleles.
Fig. 2.
Selected ion mass spectrometry to investigate the loss of lysosomal proteins in ANCL. Selected ion mass spectrometry was performed on three peptides corresponding to disease gene candidates identified by spectral counting on Man-6-P glycoproteins from patients diagnosed with ANCL (CABM-BR19 and HSB 4165) and a control case. Selected ion chromatograms are shown for a positive control protein that should be present in both samples (TPP1; Panels A and C) and CLN5 for CABM-BR19 (Panel B) and SGSH for HSB 4165 (Panel D). Arrows (Panels B and D) indicate the expected retention times for peptides from the disease gene candidates in each case.
In another ANCL case (HSB 4165), SGSH appeared to be absent by LC-MS/MS and spectral count analysis. SGSH is a lysosomal enzyme that is defective in MPSIIIA, a disorder that is unrelated to the NCLs (see above). Targeted MS was performed as described above, and failure to detect peptides corresponding to SGSH confirmed that this protein was absent in this case (Fig. 2, Panels C and D). Sequence analysis of the SGSH gene revealed that this case is a compound heterozygote for two missense mutations, E355K and S298P, both of which are documented pathogenic alleles in MPSIIIA (19, 20).
Unresolved Cases
In 14 cases, we did not identify obvious candidates for gene defects. There are various reasons why this might be the case, and these are discussed below (see “Discussion”).
Alternative Sources for the Purification of Lysosomal Man-6-P Glycoproteins
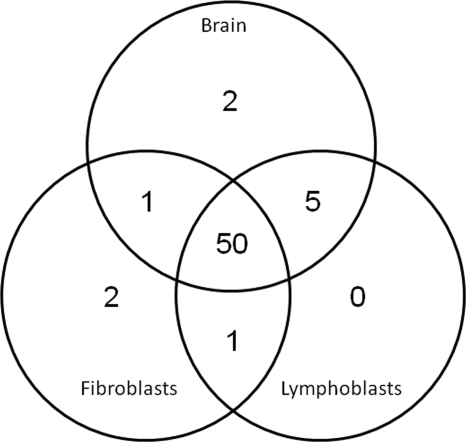
This study was designed as a preliminary investigation of the use of mass spectrometry-based proteomics methods as a tool in the identification of lysosomal disease genes. As such, there is scope for future development particularly in terms of clinical samples for analysis. Brain is highly suitable for this proteomics analysis as neurons contain high levels of phosphorylated lysosomal proteins (15), whereas in most cell types, the Man-6-P modification is rapidly removed in the lysosome by tartrate-resistant acid phosphatase (21). However, there are obvious and inherent limitations with the use of brain autopsy samples as a source for investigation, so we considered other more suitable substrates for analysis, including cultured fibroblasts and lymphoblasts. Here secretion of phosphorylated lysosomal proteins containing Man-6-P can be induced by culturing the cells in the presence of chloroquine. However, a significant concern in using material derived from sources other than the CNS to investigate these neurodegenerative diseases is whether they express the same proteome of candidate proteins that are found in brain. For both fibroblasts and lymphoblasts, we found that the range of detectable lysosomal proteins that can be purified by affinity chromatography is almost identical to brain (Fig. 3) indicating that both represent reliable samples for the proteomics investigation of neurodegenerative LSDs. Three known lysosomal proteins (cathepsin O, cathepsin K, and heparanase) that were identified in purified Man-6-P glycoprotein preparations from cell lines were not found in similar samples from brain.
Fig. 3.
Lysosomal proteins detectable in clinical samples. The distribution of lysosomal proteins identified in Man-6-P glycoprotein preparations purified from human brain (58 proteins total), immortalized lymphoblasts (56 proteins), and primary fibroblasts (54 proteins) is shown.
DISCUSSION
In this study, we used spectral counting-based protein expression profiling of purified Man-6-P glycoproteins to identify gene defects in LSD of unknown or ambiguous genetic basis. Although successful in a number of cases, this approach did not identify the gene defect in all samples examined, and this is to be expected for several reasons. First, although most defects resulting in LSDs occur within genes encoding soluble, Man-6-P-containing glycoproteins, this is not always the case. Second, some mutations may result in stable but inactive protein products, and these may also be missed. Third, proteins that are differentially expressed throughout the CNS could potentially be missed in this analysis, which was primarily focused on cerebrum. Fourth, in proteins of low relative abundance, gene defects may go unnoticed if a pathogenic loss of a protein cannot be distinguished from variability in measurement or natural biological heterogeneity. Although the analysis of rare components of mixtures is challenging, application of more sensitive, quantitative MS methods (e.g. use of isotope-labeled standards with triple quadrupole or high resolution MS) rather than spectral counting should help to diminish such errors. Regardless this proof-of-concept study for proteomics profiling using label-free methods and a linear ion trap mass spectrometer proved to be remarkably successful, identifying or confirming lysosomal defects in eight of 26 of the cases investigated (Table III).
Although most LSD alleles resulted in the loss of protein product, the example of UMB 784 highlights the possibility that some mutations may allow for significant expression levels (>1% of normal) of the mutant protein. Thus, in searching for candidates in cases where defects are not obvious, it may also be helpful to identify proteins whose expression deviates significantly from the normal. We achieved this here with a χ2 analysis (supplemental Table 5). Expression levels of some proteins (e.g. palmitoyl-protein thioesterase 1 (PPT1) and CTSD) varied widely and were frequently low even in samples with confirmed defects in other genes. However, other proteins were expressed at relatively constant levels in all samples, and a significant decrease in such proteins may be relevant. Examples include cathepsin C and α-N-acetylglucosaminidase as proteins that were significantly reduced in HSB 2356 and CABM-BR36, respectively, and these may be worth further investigation in these cases. It is also worth noting that TPP1 was significantly reduced in a case of Alexander disease (UMB 613), possibly indicating a secondary effect of disease.
An important discovery to emerge from this study was the identification of new atypical variants of unrelated lysosomal diseases that were diagnosed as ANCL. Disease course in ANCL is extremely variable, but typically onset is at about 30 years of age. It is neurodegenerative, associated with seizures and progressive dementia, and results in premature death on average 13 years after onset (for a review, see Ref. 22). The defective gene in the majority of ANCL cases remains to be identified, although several have been found to harbor mutations in PPT1 (23, 24). In this study, we investigated four ANCL cases. In one case, the protein profiling approach validated earlier genetics studies, and in another it led to the identification of the defective gene.
In an earlier study of an ANCL case of unknown etiology, CABM-BR19, we purified Man-6-P glycoproteins from a brain specimen from this patient and used two-dimensional gel electrophoresis to compare with controls, but obvious defects were not apparent (25). A genetics screen of potential disease gene candidates revealed missense changes in the gene encoding CLN5,2 a soluble lysosomal Man-6-P glycoprotein of unknown function. However, in the absence of either an activity assay for CLN5 or reliable immunoreagents, it was not clear whether these changes represented pathogenic alleles or harmless polymorphisms. Mass spectrometric profiling of purified Man-6-P glycoproteins revealed that CLN5 was absent in this case, strongly suggesting that these changes represent the cause of disease. CLN5 defects normally cause a variant form of LINCL with onset at around 4–7 years of age and survival into the second or third decade of life (18), although variants with slightly later onset (∼9 years of age) have been reported (26, 27). For the ANCL case analyzed here, onset of disease was at 20 years of age, and the patient survived until 33 years old (17). Neither of the CLN5 mutations (C126Y and Y374C)2 have been identified previously. Although Mendelian inheritance of these mutations remains to be demonstrated, the facts that CLN5 was found in all other cases except this one and that loss of CLN5 presents as an NCL disease together provide a highly convincing argument that this represents the causative defect. It is worth noting that in this study we found CLN5 to be a relatively abundant component of the mixture of proteins purified from controls when they were analyzed by LC-MS/MS, but we did not detect this protein when the same samples were fractionated by two-dimensional gel electrophoresis and the resulting spots were analyzed by either peptide mass fingerprinting or MALDI-TOF/TOF mass spectrometry (25). It is possible that CLN5 is specifically lost during the additional gel electrophoresis step or that the CLN5 peptides are not readily ionized using MALDI-based methods, but regardless, this result does demonstrate that LC-MS/MS is well suited to our current application.
SGSH defects are normally diagnosed as MPSIIIA (28) with onset that is around 2 years of age, severe neurodegeneration between 6 and 10 years of age, and survival typically into the second or third decade of life (29). Less severe SGSH alleles that result in extended lifespan have been reported (19, 20, 30–32), although onset is during childhood; thus these cases are not diagnosed as ANCL. (There is one previous report of adult onset MPSIIIA, but neurodegeneration was not a feature in this case (33).) Therefore, SGSH deficiencies have not previously been associated with ANCL. In this study, mass spectrometric protein profiling suggested that SGSH was missing in ANCL case HSB 4165, and this led to the identification of mutations in the SGSH gene. The mutations identified (E355K and S298P) are both previously described pathogenic alleles (19, 20), one of which is associated with attenuated disease (20, 32), which is consistent with our findings. It is interesting to note that although autofluorescent inclusions are not generally regarded as a defining feature of MPSIIIA they have been observed in some patients (34), which may explain possible confusion with NCLs.
These results indicate that clinically defined ANCL is not a distinct genetic entity and raise the possibility that other autosomal recessive cases actually represent late onset, slowly progressing variants of NCLs and unrelated LSDs. In support of this, an earlier review of cases published as ANCL (35) concluded that almost half either appeared atypical, had clinical features characteristic of other LSDs, or lacked a key feature of ANCL, neuronal storage. It is worth noting that both the CLN5 and SGSH-deficient cases appear to fulfill the requirements for diagnosis as ANCL outlined by Berkovic et al. (35) with electron microscopy revealing fingerprint/curvilinear profiles for the CLN5 deficiency (CABM-BR19) (17) and osmiophilic deposits for the SGSH deficiency (HSB 4165; specimen report). This indicates that the genetic origin of even bona fide ANCL is more varied than previously thought.
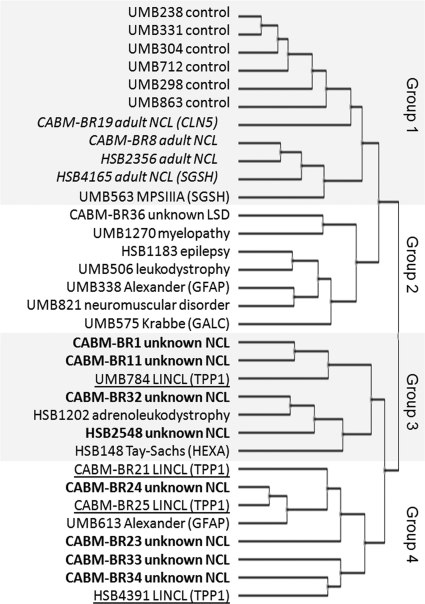
In LSDs, there are changes in lysosomal activities that are secondary to a primary defect, and these changes are likely disease-specific. Therefore we investigated the possibility that the global pattern of expression of lysosomal proteins may provide clues to the etiology of disease. In Fig. 4, we constructed a hierarchical cluster of cases based upon the expression profiles of lysosomal proteins. There are four broad groupings. In Group 1, control cases were found to be closely related, and they clustered together. All four adult NCL cases appear to be related despite differences in genetic basis (i.e. CLN5, SGSH, or unknown), and together with the case of classical MPSIIIA (SGSH), they fall within Group 1 with the controls. It is possible, for ANCL at least, that the apparent similarity to the control cases may reflect relatively subtle lysosomal alterations that result in a later onset and more slowly progressing disease than is seen in the other, more severe cases in our cohort. Group 2 comprises mainly potential LSD cases and does not include NCLs. Interestingly demyelination appears to be a common theme in most of the cases in this group. Groups 3 and 4 appear to be most different from the controls, and these largely consist of established LINCL cases and NCL cases of indeterminate origin. These groupings may provide useful clues to the identification of disease genes in cases where the mutant gene product is inactive yet stable and thus levels may not be diminished as with CTSD in ovine NCL (36). As an example, although SGSH levels were not found to be decreased, further investigation of this protein may be justified in the two unsolved ANCL cases CABM-BR8 and HSB2356 considering how closely they align with the confirmed SGSH deficiencies.
Fig. 4.
Hierarchical clustering dendrogram for normal controls and disease cases based upon relative abundance of lysosomal proteins. Normalized spectral count data (supplemental Table 2) were normalized by z-score transformation, and hierarchical cluster analysis was performed using the European Bioinformatics Institute Expression Profiler (37) using a correlation-based distance measure. Font coding indicates ANCL (indented italics), unknown NCL (bold), and LINCL (underlined).
In conclusion, the application of mass spectrometry-based methods for protein expression profiling represents a promising approach to the investigation of LSDs of unknown or ambiguous etiology. In addition to the direct identification of gene defects, the application of proteomics approaches to globally interrogate potential relationships between different diseases may also provide unique insights into both the biology of these diseases and the functions of the respective lysosomal proteins.
Acknowledgments
umerous investigators provided us with autopsy samples from unresolved cases, and their help was essential for this study. We thank Drs. Sara Mole, Jonathan Cooper, Krystyna Wisniewski, Terry Lerner, Rose-Mary Boustany, David Palmer, Aristotle Siakotos, Sidney Schochet, Hans Goebel, and David Wenger as well as Lance Johnston of the Batten Disease Support and Research Association for generous assistance in this respect. We also thank the Human Brain and Spinal Fluid Resource Center (formerly the National Neurological Research Specimen Bank) and the NICHD Brain and Tissue Bank for Developmental Disorders for additional specimens and supporting clinical information.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants DK054317 and S10RR017992 (to P. L.). This work was also supported by a grant from the Batten Disease Support and Research Association (to D. E. S.).
 The on-line version of this article (available at http://www.mcponline.org contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org contains supplemental material.
2 K. B. Sims and W. Xin, unpublished data.
1 The abbreviations used are:
- LSD
- lysosomal storage disease
- MPS
- mucopolysaccharidosis
- NCL
- neuronal ceroid lipofuscinosis
- LINCL
- late infantile neuronal ceroid lipofuscinosis
- ANCL
- adult neuronal ceroid lipofuscinosis
- Man-6-P
- mannose 6-phosphate
- MPR
- Man-6-P receptor
- SGSH
- N-sulfoglucosamine sulfohydrolase (sulfamidase)
- CTSD
- cathepsin D
- TPP1
- tripeptidyl-peptidase 1
- PPT1
- palmitoyl-protein thioesterase 1
- LTQ
- linear trap quadrupole
- GPM
- Global Proteome Machine
- CNS
- central nervous system
- OMIM
- Online Mendelian Inheritance in Man
REFERENCES
- 1.Scriver C. R.( 2001) The Metabolic and Molecular Bases of Inherited Disease, 8th Ed., McGraw-Hill, New York [Google Scholar]
- 2.McKusick V. A.( 1998) Mendelian Inheritance in Man: a Catalog of Human Genes and Genetic Disorders, 12th Ed., Johns Hopkins University Press, Baltimore [Google Scholar]
- 3.Sleat D. E., Jadot M., Lobel P.( 2007) Lysosomal proteomics and disease. Proteomics Clin. Appl. 1, 1134– 1146 [DOI] [PubMed] [Google Scholar]
- 4.Holtzman E.( 1989) Lysosomes, Plenum Press, New York [Google Scholar]
- 5.Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., Lobel P.( 2000) Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290 2298– 2301 [DOI] [PubMed] [Google Scholar]
- 6.Fan X., Zhang H., Zhang S., Bagshaw R. D., Tropak M. B., Callahan J. W., Mahuran D. J.( 2006) Identification of the gene encoding the enzyme deficient in mucopolysaccharidosis IIIC (Sanfilippo disease type C). Am. J. Hum. Genet. 79, 738– 744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sleat D. E., Donnelly R. J., Lackland H., Liu C. G., Sohar I., Pullarkat R. K., Lobel P.( 1997) Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science 277, 1802– 1805 [DOI] [PubMed] [Google Scholar]
- 8.Lübke T., Lobel P., Sleat D. E.( 2009) Proteomics of the lysosome. Biochim. Biophys. Acta 1793, 625– 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sleat D. E., Della Valle M. C., Zheng H., Moore D. F., Lobel P.( 2008) The mannose 6-phosphate glycoprotein proteome. J. Proteome Res. 7, 3010– 3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford M. M.( 1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248– 254 [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Noriega A., Grubb J. H., Talkad V., Sly W. S.( 1980) Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J. Cell Biol. 85, 839– 852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sleat D. E., Wang Y., Sohar I., Lackland H., Li Y., Li H., Zheng H., Lobel P.( 2006) Identification and validation of mannose 6-phosphate glycoproteins in human plasma reveal a wide range of lysosomal and non-lysosomal proteins. Mol. Cell. Proteomics 5, 1942– 1956 [DOI] [PubMed] [Google Scholar]
- 13.Liu H., Sadygov R. G., Yates J. R., 3rd( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193– 4201 [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., VerBerkmoes N. C., Langston M. A., Uberbacher E., Hettich R. L., Samatova N. F.( 2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5, 2909– 2918 [DOI] [PubMed] [Google Scholar]
- 15.Sleat D. E., Sohar I., Lackland H., Majercak J., Lobel P.( 1996) Rat brain contains high levels of mannose-6-phosphorylated glycoproteins including lysosomal enzymes and palmitoyl-protein thioesterase, an enzyme implicated in infantile neuronal lipofuscinosis. J. Biol. Chem. 271, 19191– 19198 [DOI] [PubMed] [Google Scholar]
- 16.Blanch L., Weber B., Guo X. H., Scott H. S., Hopwood J. J.( 1997) Molecular defects in Sanfilippo syndrome type A. Hum. Mol. Genet. 6, 787– 791 [DOI] [PubMed] [Google Scholar]
- 17.Goebel H. H., Schochet S. S., Jaynes M., Gutmann L.( 1998) Ultrastructure of the retina in adult neuronal ceroid lipofuscinosis. Acta Anat. 162, 127– 132 [DOI] [PubMed] [Google Scholar]
- 18.Savukoski M., Klockars T., Holmberg V., Santavuori P., Lander E. S., Peltonen L.( 1998) CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat. Genet. 19, 286– 288 [DOI] [PubMed] [Google Scholar]
- 19.Beesley C. E., Young E. P., Vellodi A., Winchester B. G.( 2000) Mutational analysis of Sanfilippo syndrome type A (MPS IIIA): identification of 13 novel mutations. J. Med. Genet. 37, 704– 707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunge S., Ince H., Steglich C., Kleijer W. J., Beck M., Zaremba J., van Diggelen O. P., Weber B., Hopwood J. J., Gal A.( 1997) Identification of 16 sulfamidase gene mutations including the common R74C in patients with mucopolysaccharidosis type IIIA (Sanfilippo A). Hum. Mutat. 10, 479– 485 [DOI] [PubMed] [Google Scholar]
- 21.Sun P., Sleat D. E., Lecocq M., Hayman A. R., Jadot M., Lobel P.( 2008) Acid phosphatase 5 is responsible for removing the mannose 6-phosphate recognition marker from lysosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 16590– 16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goebel H. H., Mole S. E., Lake B. D.( 1999) The Neuronal Ceroid Lipofuscinoses (Batten Disease) IOS Press; Amsterdam [Google Scholar]
- 23.van Diggelen O. P., Thobois S., Tilikete C., Zabot M. T., Keulemans J. L., van Bunderen P. A., Taschner P. E., Losekoot M., Voznyi Y. V.( 2001) Adult neuronal ceroid lipofuscinosis with palmitoyl-protein thioesterase deficiency: first adult-onset patients of a childhood disease. Ann. Neurol. 50, 269– 272 [DOI] [PubMed] [Google Scholar]
- 24.Ramadan H., Al-Din A. S., Ismail A., Balen F., Varma A., Twomey A., Watts R., Jackson M., Anderson G., Green E., Mole S. E.( 2007) Adult neuronal ceroid lipofuscinosis caused by deficiency in palmitoyl protein thioesterase 1. Neurology 68, 387– 388 [DOI] [PubMed] [Google Scholar]
- 25.Sleat D. E., Lackland H., Wang Y., Sohar I., Xiao G., Li H., Lobel P.( 2005) The human brain mannose 6-phosphate glycoproteome: a complex mixture composed of multiple isoforms of many soluble lysosomal proteins. Proteomics 5, 1520– 1532 [DOI] [PubMed] [Google Scholar]
- 26.Cannelli N., Nardocci N., Cassandrini D., Morbin M., Aiello C., Bugiani M., Criscuolo L., Zara F., Striano P., Granata T., Bertini E., Simonati A., Santorelli F. M.( 2007) Revelation of a novel CLN5 mutation in early juvenile neuronal ceroid lipofuscinosis. Neuropediatrics 38, 46– 49 [DOI] [PubMed] [Google Scholar]
- 27.Pineda-Trujillo N., Cornejo W., Carrizosa J., Wheeler R. B., Múnera S., Valencia A., Agudelo-Arango J., Cogollo A., Anderson G., Bedoya G., Mole S. E., Ruíz-Linares A.( 2005) A CLN5 mutation causing an atypical neuronal ceroid lipofuscinosis of juvenile onset. Neurology 64, 740– 742 [DOI] [PubMed] [Google Scholar]
- 28.Scott H. S., Blanch L., Guo X. H., Freeman C., Orsborn A., Baker E., Sutherland G. R., Morris C. P., Hopwood J. J.( 1995) Cloning of the sulphamidase gene and identification of mutations in Sanfilippo A syndrome. Nat. Genet. 11, 465– 467 [DOI] [PubMed] [Google Scholar]
- 29.van de Kamp J. J., Niermeijer M. F., von Figura K., Giesberts M. A.( 1981) Genetic heterogeneity and clinical variability in the Sanfilippo syndrome (types A, B, and C). Clin. Genet. 20, 152– 160 [DOI] [PubMed] [Google Scholar]
- 30.Di Natale P., Balzano N., Esposito S., Villani G. R.( 1998) Identification of molecular defects in Italian Sanfilippo A patients including 13 novel mutations. Hum. Mutat. 11, 313– 320 [DOI] [PubMed] [Google Scholar]
- 31.Gabrielli O., Coppa G. V., Bruni S., Villani G. R., Pontarelli G., Di Natale P.( 2005) An adult Sanfilippo type A patient with homozygous mutation R206P in the sulfamidase gene. Am. J. Med. Genet. A 133A, 85– 89 [DOI] [PubMed] [Google Scholar]
- 32.Meyer A., Kossow K., Gal A., Steglich C., Mühlhausen C., Ullrich K., Braulke T., Muschol N.( 2008) The mutation p.Ser298Pro in the sulphamidase gene (SGSH) is associated with a slowly progressive clinical phenotype in mucopolysaccharidosis type IIIA (Sanfilippo A syndrome). Hum. Mutat. 29, 770. [DOI] [PubMed] [Google Scholar]
- 33.Van Hove J. L., Wevers R. A., Van Cleemput J., Moerman P., Sciot R., Matthijs G., Schollen E., de Jong J. G., Carey W. F., Muller V., Nicholls C., Perkins K., Hopwood J. J.( 2003) Late-Onset visceral presentation with cardiomyopathy and without neurological symptoms of adult Sanfilippo A syndrome. Am. J. Med. Genet A 118A, 382– 387 [DOI] [PubMed] [Google Scholar]
- 34.Wisniewski K., Rudelli R., Laure-Kamionowska M., Sklower S., Houck G. E., Jr., Kieras F., Ramos P., Wisniewski H. M., Braak H.( 1985) Sanfilippo disease, type A with some features of ceroid lipofuscinosis. Neuropediatrics 16, 98– 105 [DOI] [PubMed] [Google Scholar]
- 35.Berkovic S. F., Carpenter S., Andermann F., Andermann E., Wolfe L. S.( 1988) Kufs' disease: a critical reappraisal. Brain 111, 27– 62 [DOI] [PubMed] [Google Scholar]
- 36.Tyynelä J., Sohar I., Sleat D. E., Gin R. M., Donnelly R. J., Baumann M., Haltia M., Lobel P.( 2000) A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J. 19, 2786– 2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapushesky M., Kemmeren P., Culhane A. C., Durinck S., Ihmels J., Körner C., Kull M., Torrente A., Sarkans U., Vilo J., Brazma A.( 2004) Expression Profiler: next generation: an online platform for analysis of microarray data. Nucleic Acids Res. 32, W465– 470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig R., Cortens J. P., Beavis R. C.( 2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234– 1242 [DOI] [PubMed] [Google Scholar]
- 39.Wilson E. B.( 1927) Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 22, 209– 212 [Google Scholar]