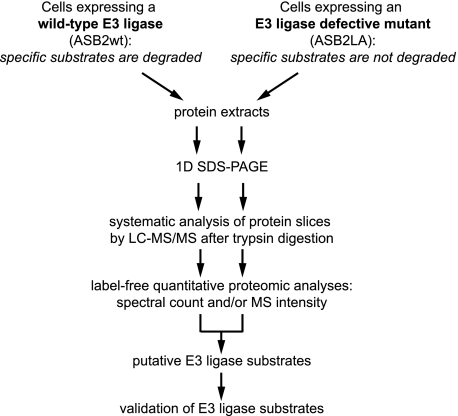
Fig. 1.
Experimental design for identification of E3 ubiquitin ligase substrates that are targeted for degradation. Our strategy uses label-free quantitative proteomics approaches to identify proteins that are absent or less abundant in cells that express a functional E3 ubiquitin ligase but that accumulate in cells that express an E3 ligase-defective mutant of this protein. This strategy was applied to identify substrates of ASB2 using myeloid leukemia cells expressing ASB2wt or an ASB2 BC box mutant (ASB2LA) that is unable to interact with the Elongin BC complex and defective in ubiquitin ligase activity under the control of an inducible promoter. 1D, one-dimensional.