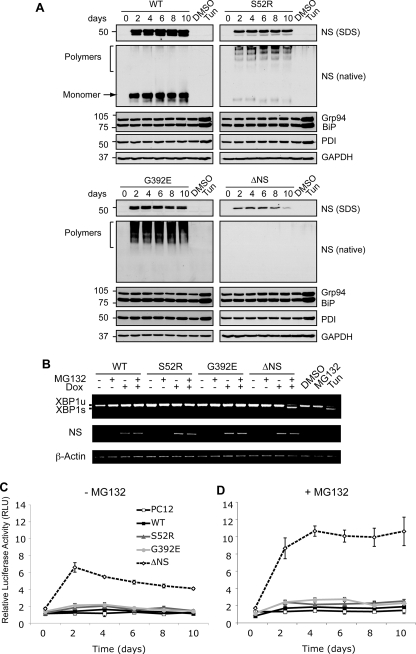
FIGURE 2.
Accumulation of neuroserpin polymers within the ER does not activate the UPR. A, polymers of neuroserpin do not up-regulate ER luminal chaperones. Neuroserpin expression was induced in PC12 cells with 10 μg/ml doxycycline for 10 days, and cell lysates were resolved by 10% w/v SDS- and 7.5% w/v/non-denaturing PAGE. Western blot analysis for neuroserpin of SDS-PAGE (NS, SDS) revealed a 50-kDa band in WT, S52R, and G392E neuroserpin-expressing cells and 28 kDa in ΔNS neuroserpin cells. On non-denaturing PAGE (NS, native), WT neuroserpin migrated as a single monomer band (arrow), whereas S52R and G392E neuroserpin formed high molecular mass ladders that are characteristic of polymers (bracket). There was no detectable signal for ΔNS on non-denaturing PAGE. The expression levels of ER luminal chaperones regulated by the UPR (glucose regulated protein 94 (Grp94), immunoglobulin heavy chain-binding protein (BiP), and protein disulfide isomerase (PDI)) were determined in the same membranes by Western blotting. Treatment with 2 μg/ml tunicamycin (Tun) for 16 h compared with vehicle alone (DMSO) was used as a positive control for UPR induction, and protein loading was assessed by Western blot analysis for GAPDH. B, polymers of neuroserpin do not activate IRE1. The expression of WT, S52R, G392E, and ΔNS neuroserpin was induced for 4 days with 10 μg/ml doxycycline (Dox), and cells were either harvested or treated 16 h prior to RNA isolation with 100 nm MG132, a reversible inhibitor of the proteasome (46). XBP1 mRNA was amplified by PCR and resolved by 2% w/v TBE agarose gel electrophoresis. Unspliced XBP1 (XBP1u) and spliced XBP1 (XBP1s) products migrated at 486 and 457 bp, respectively. Neuroserpin (NS) and β-actin cDNAs were amplified to demonstrate transgene induction and as loading controls, respectively. PC12 cells treated with 2 μg/ml tunicamycin (Tun) were used as a positive control for UPR-induced XBP1 mRNA splicing. C and D, polymers of neuroserpin do not activate ATF6. Neuroserpin expression was induced in PC12 cell lines with 10 μg/ml doxycycline for 10 days. Twenty-four hours prior to lysis the cells were co-transfected with a plasmid encoding firefly luciferase under the control of a UPR response element (p5×ATF6-Luc) and with the transfection efficiency reporter pRL-TK Renilla luciferase. The graphs show firefly luciferase normalized to Renilla luciferase as averages ± S.D. of three repeats, and values are expressed in relative light units (RLU). Cells were mock-treated (C) or treated with 100 nm MG132 (D) for 16 h prior to harvesting.