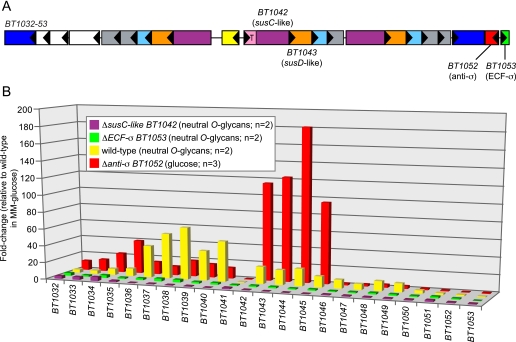
FIGURE 3.
Molecular genetic analysis of trans-envelope signaling. A, schematic of the BT1032–53 PUL, which is induced in response to mucin O-glycans. Key genes that were subjected to mutational analysis are indicated with locus tag numbers, and all genes are labeled according to the key provided in Fig. 1; white boxes in this schematic represent genes with annotated functions other than those indicated in Fig. 1. B, a histogram showing the responses of 22 genes contained within the BT1032–53 PUL to different growth and/or bacterial genetic backgrounds. For wild-type B. thetaiotaomicron grown in a preparation of neutral O-glycans (14), most genes in this PUL show increased expression relative to a MM-glucose reference (yellow bars). Individual disruptions of genes encoding either the signal transducing SusC-like protein (BT1042, purple bars) or the ECF-σ associated with this system (BT1053, green bars) result in nearly complete loss of PUL activation during growth in the neutral O-glycan mixture. Conversely, de-repression of the trans-envelope signaling switch via anti-σ mutation (red bars) results in PUL expression during growth on glucose as a sole carbon source, which does not ordinarily cause induction of these genes. Values are based on normalized GeneChip expression data. Bars indicate -fold changes in expression relative to duplicate reference datasets derived from 5-ml cultures grown in MM-glucose. All bars represent the average response of each gene in two replicate datasets.