Abstract
Lethal factor (LF), a zinc-dependent protease of high specificity produced by Bacillus anthracis, is the effector component of the binary toxin that causes death in anthrax. New therapeutics targeting the toxin are required to reduce systemic anthrax-related fatalities. In particular, new insights into the LF catalytic mechanism will be useful for the development of LF inhibitors. We evaluated the minimal length required for formation of bona fide LF substrates using substrate phage display. Phage-based selection yielded a substrate that is cleaved seven times more efficiently by LF than the peptide targeted in the protein kinase MKK6. Site-directed mutagenesis within the metal-binding site in the LF active center and within phage-selected substrates revealed a complex pattern of LF-substrate interactions. The elementary steps of LF-mediated proteolysis were resolved by the stopped-flow technique. Pre-steady-state kinetics of LF proteolysis followed a four-step mechanism as follows: initial substrate binding, rearrangement of the enzyme-substrate complex, a rate-limiting cleavage step, and product release. Examination of LF interactions with metal ions revealed an unexpected activation of the protease by Ca2+ and Mn2+. Based on the available structural and kinetic data, we propose a model for LF-substrate interaction. Resolution of the kinetic and structural parameters governing LF activity may be exploited to design new LF inhibitors.
Anthrax is an infectious disease caused by the encapsulated, spore-forming bacterium Bacillus anthracis. Systemic forms of the disease, such as inhalational anthrax, are characterized by nonspecific early symptoms, rapid progression, and lethality approaching 100% (1). The lethality of inhalational anthrax is high even with antibiotic treatment and is caused by accumulation of secreted anthrax toxin (2), which consists of three proteins as follows: protective antigen (PA),2 lethal factor (LF), and edema factor. PA binds to membrane receptors, forms pore complexes, and translocates LF and edema factor into the host cell (3, 4). The PA·LF complex is known as the lethal toxin, a virulence factor with pleiotropic action that facilitates establishment of the B. anthracis infection. LF is a Zn2+-dependent metalloprotease related to the thermolysin family that cleaves mitogen-activated protein kinase kinases (5).
Although the complete mechanism by which LF causes fatal intoxication is still unclear, inhibition of LF proteolytic activity may be an efficient means of preventing anthrax lethality. A better understanding of the LF catalytic mechanism will facilitate rational design and optimization of LF inhibitors with potential clinical applicability. Recent structural (6, 7), mechanistic (8), and in vivo studies (9, 10) of LF point to a sophisticated catalytic mechanism involving accurate recognition of multiple target substrates.
Here we use substrate phage display and stopped-flow fluorimetry kinetics to examine both the substrate specificity and elementary steps of substrate processing by LF. Our data allow us to construct a working model of LF-substrate binding and cleavage.
EXPERIMENTAL PROCEDURES
Chemicals, Materials, Bacterial Strains, and Vector DNA
Unless stated otherwise, chemicals were purchased from Sigma. The pET-22b vector and Escherichia coli strains Bl21 and BL21(DE3) were obtained from Novagen; DH12S E. coli was from Invitrogen, and pQE30 DNA was from Qiagen (Germany). Fd-tet DOG1 bacteriophage DNA was kindly provided by Dr. John P. McCafferty (Department of Biochemistry, University of Cambridge, Cambridge, UK). All solutions used in this study were made using 18-megohm ultrapure water from a Millipore synthesis station. Buffer A (30 mm Tris, pH 7.4, and 150 mm NaCl) was used as loading and washing buffer for immobilized metal affinity chromatography. Unless indicated otherwise, all other experiments were carried out in reaction buffer B (30 mm Tris-HCl, pH 7.4, and 70 mm NaCl).
Cloning, Expression, and Purification of Anthrax Lethal Factor
Full-length LF amplified from the B. anthracis Sterne strain using LFfor and LFrev primers was cloned into the BamHI and XhoI sites of a modified pET22b vector (for all primer sequences see supplemental Table I). This resulting pET-LF cytoplasmic expression construct contains N-terminal c-myc (underlined) and His6 (boldface) epitope tags (MASMTEDLEQKLISEEDLEDPHHHHHHGGSEDP) to facilitate detection and purification of target protein. E687D and H690A LF mutant constructs were generated from pET-LF using a previously described mutagenesis technique (11). The supplemental Table I contains complete list of oligonucleotides prepared for this study, and the peptides were constructed using these oligonucleotides.
Recombinant wild-type and mutant LF proteins were expressed in BL21 (DE3) E. coli cells. Cells were lysed by Triton X-100 according to a standard protocol (12), with EDTA-free inhibitor mixture (Roche Applied Science) added. Lysates were clarified by centrifugation, and LF was purified by two successive chromatographic steps using immobilized metal affinity chromatography in buffer A (Talon, Clontech) and size-exclusion chromatography in buffer B (Superdex 200 column, Amersham Biosciences). Fractions containing the expected molecular weight band by SDS-PAGE were pooled and stored at −70 °C. The protein purified according to the above procedure was electrophoretically homogeneous.
Apoenzyme was then prepared by exhaustive dialysis of LF against buffer B containing 1 mm o-phenanthroline and 10 mm EDTA, followed by reverse dialysis (two changes) against buffer B. The typical dilution factor after reverse dialysis was 500, so final concentrations of metal chelating agents were 0.004 μm for o-phenanthroline and 0.04 μm for EDTA.
Construction, Expression, and Purification of Protein FRET Substrates
Protein FRET probes were constructed using the pQE30 vector, which includes His6-tagged PS-cyan fluorescent protein 2 (PS-CFP2) and phi yellow fluorescent protein (phiYFP) as a FRET pair, as described previously (13). The LF substrates were inserted between PS-CFP2 and phiYFP as 16-amino acid linkers by cloning annealed, complementary oligonucleotides with overhangs into restriction sites SacI and PstI, located between the two genes.
The protein FRET fusion constructs were electroporated into BL21 E. coli cells, which were grown in 2× YT medium at 37 °C until the A600 reached 0.6. The temperature was then lowered to 25 °C, and cells were grown for 24 h to facilitate maturation of fluorescent protein chromophores. Fusion proteins were extracted and purified as described above for LF, except Superdex 75 (Amersham Biosciences) was used for size-exclusion chromatography. Purified monomeric fusion proteins were harvested and stored at 4 °C for up to a month with added NaN3 (0.05% v/v). Fluorescence spectra of FRET fusion proteins were verified for integrity, and FRET efficiency was analyzed using a Cary Eclipse spectrofluorimeter (Varian) with λex = 405 nm (absorption maximum of PS-CFP2).
Construction of Phage Display Libraries and Selection of Clones Cleaved by LF
Unless stated otherwise, the inserts for vector modification and library construction were prepared by annealing the indicated oligonucleotides (see supplemental Table I for oligonucleotide and peptide sequences), filling-in by Klenow fragment, and digesting with appropriate restriction enzymes.
The phage vector Fd-tet DOG1 was modified to prepare the Fd-Base vector used for all further library construction work. A 623-bp fragment amplified using XhoFdFor and BamFdRev primers was cut by BamHI and PstI and cloned into the BamHI and PstI sites of Fd-tet DOG1. The TAA stop codon was introduced immediately upstream of the XhoI site to prevent propagation of phages lacking library inserts.
Positive control phage was prepared using annealed FdMycFor and +SuPhageRev oligonucleotides digested with ApaLI and XhoI and cloned into the ApaLI and XhoI sites of FdBase. The N terminus of mature pIII was AQTEQKLISEEDLGGSGRRKKVYPYPMELE, where c-myc is italicized, and substrate (LF15) is underlined (see supplemental Table I and Ref. 7 for details), and the linker is shown in boldface.
Negative control phage was prepared similarly using FdMycFor and −SuPhageRev oligonucleotides to generate the MAQTEQKLISEEDLGGSGRLE N terminus of mature pIII, with a single arginine introduced to facilitate trypsin cleavage.
The phage library was constructed by cloning annealed FdMycFor and RandFdXho oligonucleotides into FdBase. 5-Fold molar excess of the duplex digested by ApaLI and XhoI was ligated with 40 μg of ApaLI- and XhoI-digested FdBase DNA. Electroporation of ligated DNA into E. coli DH12S yielded ∼109 individual transformants. Fifty clones were randomly chosen for verification of the insert by sequencing. The generic structure of the N terminus of the mature pIII displaying the random peptide library was AQTEQKLISEEDLGGSGNNNNNNNNSLE.
To prepare the second-iteration library, first an insert containing the double c-myc tag, the extended linker, and the LF target peptide was prepared from 2MycAscFor and 2MycAscRev oligonucleotides digested by ApaLI and XhoI and ligated into ApaLI- and XhoI-digested Fd-Base. This modification introduced an AscI site into the vector for subsequent library construction. Positive and negative control phage were prepared using the oligonucleotides LibAscFor and +SuPhageRev or −SuPhageRev.
The insert for the second-iteration library was prepared from LibAscFor and 2LibRev and cloned into AscI-XhoI of the modified FdBase plasmid to yield the second-iteration library, in which the N terminus of mature pIII is AQTEQKLISEEDLSGSGEQKLISEEDLGSGAPSGSDLGGSGRX(T/S/F)RR(V/I)(T/S/N/K/P/H/I)XXXXXLE. The complexity (∼108 individual clones) and integrity of the second-iteration library were verified as described above. Libraries were plated with 0.6% agarose (100 ml) in 2× YT and tetracycline (10 μg/ml) on a 1.5% 2× YT agar layer with 10 μg/ml tetracycline. After overnight growth, the agarose layer was scraped, mixed with 50 ml of 2× YT with 50% ultrapure glycerol (Invitrogen), and homogenized. The homogenate was distributed in 10-ml aliquots and stored at −80 °C until use.
The library aliquot was diluted in 500 ml of 2× YT with 10 μg/ml tetracycline and incubated at 25 °C for 12 h. After removal of cells and debris by two sequential 10-min spins at 10,000 × g at 4 °C, phage particles were isolated by double precipitation with 1/5 volume of ice-cold 20% polyethylene glycol 8000, 2.5 m NaCl. An aliquot of 1011 purified phage particles was mixed with 50 μg of 9E10 antibody (ATCC CRL-1729) in 100 μl of buffer B, incubated at 4 °C for 3 h, and captured by Pansorbin (Calbiochem), following the manufacturer's instructions.
After extensive washing (40 times in buffer B with 0.05% Tween 20), immobilized phage were treated with 100 nm LF for 4 h at 37 °C in buffer B (100 μl). Released phage were harvested, titrated, and used to infect E. coli. Infected cells were plated in 30 ml of 0.6% agarose, 2× YT medium with 10 μg/ml tetracycline and cultured at 37 °C overnight. Infected cells were collected as described above and used for subsequent rounds of screening.
Positive and negative control phage treated under the same conditions were analyzed after being plated onto 1.5% 2× YT agar plates with 10 μg/ml tetracycline. The presence of sequences encoding substrates of the correct length in isolated clones was verified by PCR after each selection round.
Phage ELISA
Individual phage clones were screened for LF cleavage by ELISA in standard 96-well microtiter plates (Nunc Maxisorp). Briefly, 1011 purified phages in phosphate-buffered saline (1.7 mm NaH2PO4, 5.2 mm Na2HPO4, and 150 mm NaCl, pH 7.4) were applied to each well and incubated for 1 h at 25 °C. The wells were then washed three times in phosphate-buffered saline, 0.05% Tween 20, blocked with 1% bovine serum albumin, and incubated with LF in test wells and without LF in control wells. Then the wells were washed three times, incubated with 9E10 antibody (0.1 μg/ml) for 1 h, then with anti-Fc antibody (1:40,000 dilution; Amersham Biosciences) for 1 h, and developed with o-phenylenediamine (Sigma) according to the manufacturer's instructions. ELISA readouts were performed with a TECAN GeNios microplate fluorimeter. Clones with peptides substrates susceptible to LF cleavage were identified by comparison between control and test wells.
Analysis of LF Cleavage of Protein FRET Substrates under Steady-state Conditions
LF activity was measured using protein FRET substrates in buffer B with 10 μm CaCl2 (except all experiments with apo-LF) to prevent precipitation of protein FRET substrates. Reaction progression was monitored by FRET loss using 96-well black microtiter plates (Greiner, Austria) and a TECAN GeNios microplate reader (excitation, 405 nm; emission, 535 nm). The concentration of LF15P (LF15-based fluorescent protein substrate (7)) ranged between 0.5 and 20 μm. In all other experiments, 0.5 μm of FRET substrate and excess LF (2.5–25 μm) were used.
Collection of steady-state kinetic data for apo-LF activation by divalent metal ions was performed at 20 nm of apo-LF and 0.5–5 μm of LF15P. The concentration of metal ions (Zn2+, Ca2+ and Mn2+) was 50 μm. Kinetic data were analyzed and fitted using linear regression by Sigma Plot Enzyme Kinetics Module version 1.1 software (SSPS Inc.).
Peptide Synthesis and Cleavage Site Determination
Fluorescein or dansyl-labeled peptide substrates for LF were prepared using a Syro I synthesizer (MultiSynTech, Germany) using Fmoc/t-butyl chemistry. The identity of synthetic peptides was confirmed by matrix-assisted laser desorption ionization-mass spectrometry, and the purity was assayed by analytical reverse-phase HPLC (>95%). The position of the scissile bond was determined by mass spectrometry. Specifically, the peptide cleavage products were separated by HPLC, and the molecular weight of the dye-labeled product was verified using mass spectrometry. The sequences of all synthesized peptides are shown in supplemental Table I.
HPLC Analysis
Stock solution of LF15F (LF15-based peptide substrate (7) N-terminally labeled with fluorescein) (1 mm in dimethyl formamide) was stored at −20 °C and protected from light. The substrate (1–50 μm) was incubated with 50 nm LF in buffer B for 0–420 s. HPLC analysis was performed as described previously (14). The reaction was stopped with 10-fold v/v excess of 60% CH3CN in water to precipitate LF. The samples were then vortexed, allowed to sit on ice for at least 15 min, and centrifuged (2,000 × g for 60 s), and the pellet was discarded. The substrate and product were separated on a Symmetry 300 C18 5-μm 3.9 × 150-mm HPLC column (Waters) with a CH3CN/H2O gradient (with 0.1% trifluoroacetic acid) on a Waters 501 HPLC system with fluorescence detection (Waters 470 scanning fluorescent detector).
Pre-steady-state Kinetics, Data Collection, and Analysis
Stopped-flow measurements were carried out using a model SX.18MV stopped-flow spectrofluorimeter (Applied Photophysics). LF15F was excited at λex = 495 nm, and emission was detected using a long pass Schott filter WG530 (Schott, Mainz, Germany) at λem >530 nm. For detection of FRET loss after cleavage of the protein FRET substrate LF15P, the PS-CFP2 FRET donor was excited at 405 nm, and emission of the phiYFP FRET acceptor was detected at 530 nm. In the FRET-based detection of dansylated peptide substrates (LF15D and S20D), tryptophan fluorescence of LF was excited at 280 nm, and dansyl acceptor emission was measured at 560 nm. The dead time of the instrument was 1.4 ms. Typically, the substrate concentration was fixed, and the enzyme concentration was varied within 2 orders of magnitude of the substrate concentration. In all experiments, substrate concentrations in the reaction chamber after mixing were as follows: LF15F, 5 μm; LF15D, 5 μm; S20D, 4 μm; LF15P, 0.25 μm. LF inhibition by In-2-LF hydroxamate (GYβARRRRRRRRVLR-NHOH; Calbiochem) was carried out using fixed concentrations of LF15F (5 μm) and LF (0.5 μm) and a range of inhibitor concentrations (0.1–2.5 μm).
In the pre-steady-state experiments, apo-LF was activated by rapidly mixing the protease with solution containing the substrate (LF15P) and divalent ions in the reaction vessel of the stopped-flow device. The substrate concentration was 0.25 μm; the LF concentration was 0.25 μm, and the concentration of metal ions ranged between 0.5 and 250 μm for Zn2+, between 10 and 1000 μm for Ca2+, and between 10 and 250 μm for Mn2+ and Mg2+.
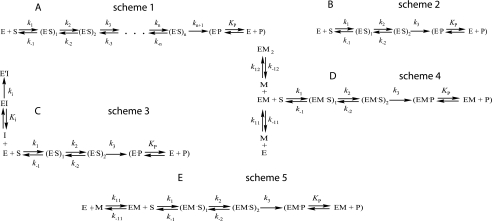
The kinetic data were obtained by numerical fitting using DynaFit software (BioKin, Pullman, WA) (15), as described previously (16, 17). Data are presented as the mean of at least three independent experiments. Kinetic schemes were built as sequences of n reversible and one irreversible steps, and an enzyme·product complex equilibrium stage (Fig. 2A, scheme 1). The conformity of the fitted kinetic models to the experimental data was assessed by monitoring residuals against time for different scheme fits in addition to using the scree test (17). Typically, this was achieved by following the dependence of standard deviations of the data residuals after fitting to an n-step binding model versus n. A smooth decrease in the standard deviation that leveled off after a certain number (N) of steps, indicated that the N-step kinetic scheme was indeed the minimal one (see supplemental Fig. 2 and under “Results” for further explanation).
FIGURE 2.
Kinetic schemes fitted to the experimental data. E, enzyme; S, substrate; P, product. A, simulation of N-step substrate binding mechanism. B, simulation of two-step reversible binding and one-step irreversible cleavage step mechanism used for characterization of the LF-substrate interaction. C, simulated mechanism of LF inhibition by peptide hydroxamate In-2-LF; I, inhibitor. D, mechanism of LF apoenzyme activation by Zn2+; M, metal ion (Zn2+). E, mechanism of LF apoenzyme activation by Ca2+ or Mn2+; M, metal ion (Ca2+ or Mn2+). Fitting was done with DynaFit software (15). To verify pre-steady-state kinetic data, the scree statistical test (17) was used for each mechanism (see supplemental Fig. 2 for details).
RESULTS
Phage Display Library Screening and Substrate Selection
The conserved portion of the LF substrate in MKKs consists of only eight amino acids (18), and even seven-amino acid sequences can serve as LF substrates (8). However, longer substrates can be cleaved more efficiently because of secondary interactions between LF and its substrate (10). We used phage display screening to determine the minimal length of substrate efficiently cleaved by LF and to identify substrate determinants responsible for optimal subsite occupancy in the LF-substrate interaction (19).
The library of peptide substrates was displayed on the surface of the Fd phage. This results in polyvalent display of randomized eight-amino acid peptides inserted near the N terminus of the full-length minor phage coat protein pIII. The randomized sequences were programmed by NNV codons, where N is A, G, C, or T and V is G, C, or T. The c-myc bait recognized by the 9E10 mAb was placed at the N terminus of pIII and separated from the randomized region with a flexible GGSG linker (supplemental Table I).
Screening was performed by immobilizing the phage-displayed library on a solid phase (Pansorbin) via 9E10 mAb followed by addition of LF. Phage elution should only occur for phage particles cleaved by LF and not for those carrying aberrant c-myc tags and thus escaping capture by the 9E10 mAb if treatment by LF is carried out in solution.
To prove that c-myc-bound 9E10 mAb does not block LF access to phage-displayed substrate, positive (+) and negative (−) control phages were prepared (supplemental Table I). The positive control phage had essentially the same pIII structure as the library phage, except it displayed LF15 peptide (RRKKVYPYPME (7)), whereas the negative control phage contained single arginine instead of a displayed peptide and lacked any LF substrate-like sequences. Cleavage by trypsin eluted 100% of Pansorbin-attached (+)-phages and 50% of (−)-phages in 2 h. Cleavage by LF for 2 h removed up to 70% of (+)-phages and less than 0.1% of (−)-phages. The 2–5-h incubation in reaction buffer without protease released less than 0.1% of either phage type. Thus, attachment of phage to Pansorbin via c-myc/9E10 binding does not block access of LF to the scissile bond of a displayed substrate.
After 10 rounds of selection, 50 randomly picked clones were assayed using phage ELISA. Sequences of nine clones cleaved by LF according to the ELISA data are presented in supplemental Table II. Three substrates were cleaved in protein FRET assay, where LF-sensitive peptides were used as linkers between the two fluorescent proteins, PS-CFP2 and phiYFP. After cleavage within the linker, the fluorescent proteins become physically separated resulting in a loss of FRET.
Cleavage efficiency of these substrates was low, with protein FRET substrates showing detectable conversion after 18 h of incubation with LF. Cleavage was also confirmed by electrophoresis of fusion proteins treated with LF (data not shown). Alignment of selected clones against the LF15 substrate (7) and peptide derived from MKK6 (supplemental Table II) revealed that in the majority of substrates, including the LF15 peptide and a family of artificially designed LF substrates (8, 20), the central region (p5 to p3 positions) contains a BBφ motif (where B is a basic amino acid (R or K), φ is a hydrophobic amino acid, most frequently V or I). This suggests that invariant amino acids of the pIII protein were recruited to form the scissile bond.
The presence of invariant amino acids in the selected substrates suggests that the length of the randomized peptide must be increased to obtain substrates with optimal subsite occupancy. We therefore prepared a secondary library in which the length of the substrate was increased to 12 amino acid residues. Conserved residues identified in the first screen were fixed. Some diversity was introduced at the semi-conserved positions, whereas positions P1 to P4′ were randomized using NNV codons, as described above (Table 1). A double c-myc tag was incorporated upstream of the substrate sequences to increase the anchoring efficiency of the phage on the solid phase (21), thus decreasing the likelihood of selecting spontaneously dissociated phage particles instead of LF cleavage products. The length of flexible spacer between the substrate and bait was increased to 14 amino acid residues.
TABLE 1.
LF substrates isolated from second-iteration substrate phage library
Positions surrounding the scissile bond are numbered as P8 to P4′ from the N to C terminus of the substrate. The scissile bond is P1 to P1′. Substrates were selected as cleavable by LF after screening the second-iteration library. To prepare the second-iteration library, conserved amino acids identified from the eight-amino acid library screening were fixed, and other positions were either partially (P7, P6, P3, and P2) or fully (P1 to P4′) randomized. Values accompanying amino acid symbols reflect the frequency of respective amino acids (%) at the indicated positions calculated after analysis of 31 isolated LF substrates. Only amino acid residues with ≥4% frequency are shown. N, any nucleotide; V, C/T/G; W, A/T; Y, C/T; R, G/A; D, A/G/T; H, A/C/T; X, any amino acid. Several abundant amino acids (for example, F in p3) were not programmed and may have emerged by spontaneous point mutation. LF15 is a substrate that was previously isolated from a synthetic peptide library (7) and is cleaved efficiently by LF.
| Cleavage positions |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | |
| Sub-library DNA sequence | CGT | DWH | WYC | CGT | CGC | RTT | WHH | NNV | NNV | NNV | NNV | NNV |
| Sub-library amino acid sequence | R | K/N/I/E/D/V/Y/L/F | T/S/I/F | R | R | V/I | T/S/N/K/P/H/I | X | X | X | X | X |
| Frequency of amino acid residues, % | R(100) | I(60.46) | I(63.04) | R(100) | R(100) | V(82.61) | N(47.83) | S(47.83) | S(56.52) | L(51.16) | P(47.73) | L(63.04) |
| D(11.63) | F(19.57) | I(13.04) | S(13.04) | A(10.87) | P(13.04) | R(6.98) | L(15.91) | G(6.52) | ||||
| Y(9.30) | R(8.70) | F(4.35) | I(10.87) | L(8.70) | T(6.52) | G(6.98) | I(6.82) | H(6.52) | ||||
| V(6.98) | T(4.35) | L(8.70) | P(6.52) | G(4.35) | S(6.98) | G(4.55) | R(6.52) | |||||
| E(4.65) | S(4.35) | T(8.70) | R(6.52) | Y(4.35) | K(4.65) | S(4.55) | E(4.35) | |||||
| N(4.65) | E(4.35) | I(4.65) | V(4.35) | |||||||||
| G(4.35) | P(4.65) | |||||||||||
| The resulting consensus | R | I | I | R | R | V | N | S | S | L | P | L |
| LF15 | R | R | K | K | V | Y | P | Y | P | M | E | |
The resulting phage library was screened as described above for five rounds. Fifty random clones were then picked and assayed by phage ELISA; of these, 31 were sequenced. The frequency of amino acid residues in each position is shown in Table 1. The most abundant substrate was consensus S82 (15 of 31 clones), but it was not the most efficiently cleaved substrate (Table 2). Cleavage of selected substrates was further studied using protein FRET. A peptide encompassing the LF cleavage site in the MKK6 kinase and the LF15 peptide (7) were used as controls.
TABLE 2.
Structural and steady-state kinetic data for native and mutant LF substrates selected from a phage library
Kinetic parameters for substrates were obtained using protein FRET. Peptides were engineered as linkers between fluorescent proteins. Cleavage of the linker separated the FRET pair members PSCFP2 and phiYFP, resulting in loss of FRET signal and enabling fluorometric analysis of the reaction progression. Amino acid sequences of peptide substrates are numbered P8 to P4′ from the N to C terminus. S indicates substrates were isolated from the phage library. LF15P is the protein FRET substrate constructed based on the LF15 peptide (7). MKK6P was prepared using the LF cleavage site in Mkk6 kinase (18). Amino acid positions subjected to site-directed mutagenesis are underlined. Kinetic data were collected in triplicate and fitted using Sigma Plot enzyme Kinetics software.
| Mut no. | Description of substrate | Peptide substrate composition |
Kinetic parameters |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | Km·10−6 | kcat·10−4 | kcat/Km | ||
| m | s−1 | m−1 s−1 | ||||||||||||||
| LF15P | K | R | R | K | K | V | Y | P | Y | P | M | E | 5.4 ± 0.5 | (5 ± 0.3)·103 | 92593 | |
| MKK6P | K | K | R | N | P | G | L | K | I | P | K | E | 29.0 ± 3.7 | 6.1 ± 1.2 | 21 | |
| S82 | R | I | I | R | R | V | N | S | S | L | P | L | 15.7 ± 2.3 | 0.9 ± 0.2 | 6 | |
| S40 | R | D | F | R | R | I | I | A | E | R | Y | L | 6.6 ± 1.3 | 1.9 ± 0.3 | 28 | |
| S20 | R | D | I | R | R | I | T | L | F | S | L | H | 3.4 ± 0.7 | 5.0 ± 0.5 | 147 | |
| 1 | S20 Lp1P | R | D | I | R | R | I | T | P | F | S | L | H | 17.1 ± 4.2 | 2.1 ± 0.4 | 12 |
| 2 | S20 Fp1′Y | R | D | I | R | R | I | T | L | Y | S | L | H | 2.1 ± 0.7 | 0.8 ± 0.2 | 38 |
| 3 | S20 Ip3V/Tp2Y/Lp1P | R | D | I | R | R | V | Y | P | F | S | L | H | 4.2 ± 1.2 | 0.9 ± 0.1 | 22.7 |
| 4 | S20 Dp7K/Ip6K | R | K | K | R | R | I | T | L | F | S | L | H | 86.8 ± 10.2 | 3.93 ± 0.8 | 4.53 |
| 5 | S20 Dp7K/Ip6K/Ip3V/Tp2Y | R | K | K | R | R | V | Y | L | F | S | L | H | 1.0 ± 0.1 | 9.8 ± 0.3 | 946 |
| 6 | S20 Dp7K/Ip3V/Tp2Y/Lp1P/Sp2′P | R | K | I | R | R | V | Y | P | F | P | L | H | 5.9 ± 2.0 | 116.0 ± 33.7 | 1960 |
Validation of the protein FRET system to analyze steady-state kinetics of substrate cleavage was performed by comparing kinetic data for the fluorescein-labeled LF15 peptide (LF15F),obtained by HPLC, and the protein FRET construct incorporating LF15 (LF15P) (see steady-state Km and kcat in Table 3 and supplemental Fig. 1A). The steady-state kinetic parameters were almost identical for LF15F and LF15P.
TABLE 3.
Pre-steady-state kinetic parameters for LF-mediated proteolysis
Pre-steady-state kinetic parameters for all proteolytic reactions except inhibition of LF15F cleavage by In-2-Lf were derived from data shown in Fig. 1 and were fitted to kinetic data (Fig. 2B, scheme 2). Inhibition of LF15F cleavage by In-2-LF was simulated as shown in Fig. 2C, scheme 3. All experiments were done in triplicate, and data were fitted using Dynafit software (15). Km and kcat values for pre-steady-state conditions were calculated based on equations derived from Fig. 2B, scheme 2 using enzyme kinetic graph theory (32). Constants for steady-state conditions were obtained using the Michaelis-Menten equation. LF15P, protein FRET substrate; LF15D, dansylated LF15 peptide; LF15F, fluorescein-labeled LF15 peptide; S20D, dansylated S20 peptide.
| Kinetic constants | Substrate type |
||||
|---|---|---|---|---|---|
| LF15P | LF15F | LF15D | S20D | LF15F+ In-2-LF inhibitor | |
| k1, m−1 × s−1 | (42.3 ± 6.9) × 106 | (39.1 ± 6.2) × 106 | (6.3 ± 0.3) × 106 | (0.35 ± 0.02) × 106 | (57.4 ± 2.4) × 106 |
| k−1, s−1 | 219.3 ± 10.3 | 245.7 ± 13.5 | 17.3 ± 1.6 | 7.1 ± 0.4 | 209.9 ± 7.3 |
| Kbind1, m−1 | 1.93 × 105 | 1.59 × 105 | 3.64 × 105 | 4.9 × 104 | 2.7 × 105 |
| k2, s−1 | 3.9 ± 0.2 | 3.8 ± 0.1 | 7.7 ± 0.2 | 0.15 ± 0.02 | 4.0 ± 0.1 |
| k−2, s−1 | 1.3 ± 0.1 | 1.4 ± 0.1 | 2.1 ± 0.1 | 0.22 ± 0.03 | 1.1 ± 0.4 |
| Kbind2 | 1.9 | 1.7 | 2.75 | 0.68 | 3.63 |
| k3, s−1 | 0.77 ± 0.04 | 0.83 ± 0.01 | 0.7 ± 0.1 | ND | 0.8 ± 0.1 |
| Kp, m | (1.3 ± 0.1) × 10−6 | (1.3 ± 0.1) × 10−6 | (5.2 ± 0.3) × 10−6 | ND | (2.1 ± 0.4) × 10−6 |
| Km, m | |||||
| Pre-steady state | 1.8 × 10−6 | 2.3 × 10−6 | 8 × 10−7 | 1.2 × 10−5 | 1.2 × 10−6 |
| Steady state | (5.4 ± 0.5) × 10−6 | (6.2 ± 3.3) × 10−6 | ND | ND | ND |
| kcat s−1 | |||||
| Pre-steady state | 0.50 | 0.52 | 0.51 | 4.4 × 10−4 | 0.54 |
| Steady state | 0.5 ± 0.03 | 0.98 ± 0.22 | ND | ND | ND |
| Ki, m | (2.2 ± 0.3) × 10−8 | ||||
| ki s−1 | (9.6 ± 1.9) × 10−3 | ||||
The sequences of the most efficiently cleaved substrates, S20, S40, and S82, are shown in Table 2. Among the selected substrates, the greatest LF cleavage efficiency was observed for the S20, for which the Km value was 7 times smaller, and the kcat value was similar to that of the MKK6P derived from the LF cleavage site in MKK6 (Table 2).
Site-directed mutants of the S20 protein FRET substrate were constructed to analyze the role of various portions of LF substrate for overall cleavage susceptibility (Table 2). Mutation of the P1 leucine residue of S20 to proline, which is abundant in natural and artificial substrates (7), resulted in a 5-fold increase in Km and a 2.5-fold decrease in kcat (mutant 1 in Table 2). A conservative Phe-to-Tyr mutation (mutant 2) in the P1′ position yields a minor overall loss in cleavage efficiency (kcat/Km). Introduction of a VYP stretch instead of ITL in the p3 to p1 positions of S20 (mutant 3) resulted in a 7-fold decrease in kcat compared with the parental substrate, likely because of the Leu-to-Pro replacement similar to that introduced in mutant 1. Mutation of N terminus of the substrate by replacing of P6 and P7 with lysines (mutant 4) resulted in a significant increase of Km compared with the S20 peptide. Strikingly, replacement of P3 and P2 in mutant 4 with Val and Tyr (mutant 5), respectively, resulted in almost 100-fold decrease in Km and a more than 2-fold increase in kcat comparing to mutant 4. In mutant 6, P7 was replaced by lysine and P1 and P2′ were mutated to prolines. This resulted in a further increase in cleavage efficiency. Although Km was increased slightly compared with mutant 5, a greater than 10-fold gain in kcat was observed.
Collectively, our site-directed mutagenesis data reveal a complex interplay between changes in amino acid residues and cleavage susceptibility in the LF S20 substrate. Concerted replacement of certain amino acid groups at different parts of a substrate rather than simple point mutations are needed to increase the cleavage susceptibility.
Peptides encompassing the P8 to P4′ residues of the S20, S40, and S82 substrates were synthesized, and the scissile bond for LF cleavage was determined directly using liquid chromatography/mass spectrometry. In all cases, the position of the scissile bond coincided with the predicted one, as depicted in Table 2. No additional cleavage sites were identified. The cleavage data validated the initial assumption regarding the positions of conserved residues in phage-selected LF substrates and the specificity of LF-mediated hydrolysis.
Analysis of Pre-steady-state Kinetics of LF-mediated Proteolysis, Cleavage of LF15-based Substrates
The most effectively cleaved LF substrate, LF15 (7), was chosen for analysis of pre-steady-state kinetics of LF-mediated proteolysis. Three substrates based on this peptide were designed for analysis using different techniques to probe enzyme-substrate interactions. In the first substrate, LF15P, the peptide acts a linker between two fluorescent proteins, PS-CFP2 and phiYFP. After cleavage within the linker, the fluorescent proteins become physically separated, resulting in a loss of FRET signal. The second substrate, LF15F, was N-terminally labeled with fluorescein, whose fluorescence is quenched because of its amino acid environment. After cleavage, this environment changes, and the fluorescence intensity of LF15F increases, thus enabling easy detection of the cleavage event and product accumulation. The third peptide, LF15D, was N-terminally labeled with dansyl and was used to detect enzyme-substrate interactions by observing tryptophan-dansyl FRET signal (22).
Assays based on physical principles other than fluorescence detection were also used to provide direct demonstration of LF-mediated cleavage. Cleavage of protein FRET fusions was demonstrated using SDS-PAGE (supplemental Fig. 1, B and C). Chromatographic HPLC-based assays were used for analysis of cleavage of LF15F (supplemental Fig. 1A) and LF15D (data not shown). The pre-steady-state kinetic profiles for all three LF15-based substrates are presented in Fig. 1. At the equimolar LF/substrate ratio, cleavage was almost complete in 60 s. Thus, despite the differences in both fluorophore and the protein environment, all the substrates demonstrated similar cleavage kinetics in the pre-steady-state experiments, consistent with the similar steady-state kinetic data obtained for LF15P and LF15F.
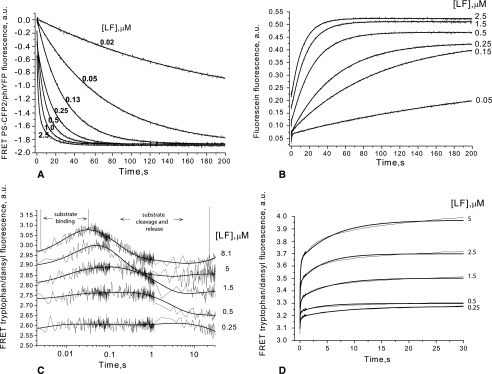
FIGURE 1.
Time course for LF-mediated hydrolysis of fluorescent substrates. The reaction was monitored using a stopped-flow spectrofluorometer. Substrates used and the readout collected were as follows. A, for LF15P (LF15 peptide placed between fluorescent proteins PS-CFP2 and phiYFP), the decrease of FRET signal between fluorescent proteins was measured. B, for the fluorescein-labeled peptide substrate LF15F, the fluorescence increase was measured during LF proteolysis. C, for the dansyl-labeled peptide substrate LF15D, FRET signal between enzyme tryptophan residues and dansyl was measured. D, tryptophan-dansyl FRET was also measured in the case of the phage-selected dansyl-labeled peptide substrate S20D. Substrate concentrations used were 0.25 μm (A), 5 μm (B and C), and 4 μm (D); LF concentrations are indicated in the panels. Fig. 2B, scheme 2, was fitted to these data (smooth lines) using DynaFit software (15). Resulting parameters are given in Table 3. To build each experimental curve, data were collected in triplicate.
The most informative data regarding the LF proteolytic mechanism was provided by cleavage of the LF15D substrate (Fig. 1C). The dansyl group of the peptide can form FRET pairs with tryptophan residues of LF (22). LF contains five tryptophan residues, of which Trp-570 and Trp-606 are known to be in close proximity to the active site (6). Based on known structures and taking into account the effective FRET distance (10–100 Å), one can infer that the energy transfer between the Trp-606 or Trp-570 of LF and LF15D should produce detectable changes in dansyl fluorescence upon binding and subsequent physical separation of the enzyme and substrate, which leads to loss of FRET signal (Fig. 1C).
The kinetic curves for the experimental data show an excellent fit (DynaFit software) (12) to data from computer simulations (Fig. 1). The best fit model is a four-step reaction mechanism (Fig. 2B, scheme 2) with two reversible steps leading to formation of the enzyme·substrate complex followed by irreversible catalysis of bond cleavage and rapid reversible dissociation of the product (Fig. 2B).
Validation of the reaction model was performed using scree test (17). The standard deviation decreases and levels off after one step, indicating that the two-step kinetic scheme is indeed the minimal one for LF binding to LF15F at a 1:1 molar ratio (supplemental Fig. 2). Application of the scree test to kinetic schemes for LF15P and LF15D cleavage was also consistent with a two-step mechanism for LF-substrate binding. Rate and equilibrium constants for proteolysis of LF15-based substrates are presented in Table 3. Values of elementary constants vary only slightly between the three substrates studied. In addition, two binding steps were found for all of the substrates tested, with the rate-limiting step being peptide bond hydrolysis.
Inhibition of LF protease activity was examined using LF15F and the peptidyl hydroxamate inhibitor In-2-LF (20). In-2-LF binds to LF and chelates Zn2+ ions at the enzyme active site. The dissociation rate of Zn2+ is slow (Ki = 1 nm) and is made even slower by the binding of the peptidyl component of the inhibitor to LF. Thus, within the time frame of the stopped-flow studies, In-2-LF is an irreversible LF inhibitor (Fig. 2C, scheme 3) and therefore depicts primary reversible binding of In-2-LF to the protease with subsequent chelation of Zn2+ by the hydroxamate moiety.
Values of reaction constants for LF15F cleavage in the presence of inhibitor are shown in Table 3. These results indicate that the inhibitor binds to enzyme unoccupied by substrate, and that the elementary constants of substrate binding and cleavage reactions are similar in the presence and absence of In-2-LF. Attempts to fit the LF inhibition data to the alternative kinetic scheme, in which In-2-LF interacts with either LF·substrate complex (ES1 or ES2), failed. Thus, it is likely that the inhibitor removes free LF from the reaction rather than interfering with rearrangement of substrate-bound LF.
Hydrolysis of LF15F by LF Active Site Mutants E687D and H690A
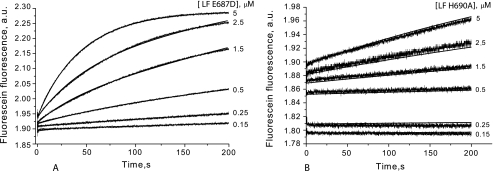
Site-directed LF mutants were tested for LF15F hydrolysis using the stopped-flow technique (Fig. 3). His-690 is one of three amino acid residues that coordinate the divalent metal ion in the LF active site, and Glu-687 likely activates the water molecule bound to Zn2+ at the active site (6). H690A and E687D mutations almost completely abolish LF activity (23). However, under pre-steady-state conditions with equimolar enzyme/substrate concentrations, we still observed some changes in substrate fluorescence intensity with the mutant forms of LF (Fig. 3, A and B). The effect was much smaller than what was observed with wild-type LF (Fig. 1B), with the H690A mutant showing the smallest change in fluorescence intensity. These results indicate that the mutant LF species have a significantly lower rate of substrate conversion.
FIGURE 3.
Cleavage of the fluorescein-labeled peptide substrate LF15F by LF E687D (A) and LF H690A (B) mutant enzymes examined by stopped flow. Fluorescence increase measured during the enzyme cleavage was used as the readout. The substrate concentration was 5 μm; enzyme concentrations are indicated to the right of the panels. The calculated kinetic parameters are shown in Table 4.
To demonstrate that the fluorescence changes caused by LF mutants are because of residual catalytic activity, we incubated LF15F with the fully inactive apo-LF. Addition of apo-LF did not cause any detectable changes in LF15F fluorescence (supplemental Fig. 4A). To prove that the fluorescence changes detected in the stopped-flow experiments (Fig. 3) are because of residual protease activity of LF mutants, we analyzed LF15F cleavage by both LF mutants and apo-LF with reverse-phase HPLC (supplemental Fig. 4B). These data confirmed the substrate hydrolysis by LF E687D and LF H690A but not apo-LF.
Rate constants for mutant LF species were calculated basing on the sequence of the elementary reaction steps presented in Fig. 2B and were compared with those calculated for the wild-type enzyme (Table 4). Both mutations decreased the efficiency of ES1 formation (k1 and k−1) and peptide bond cleavage (k3), whereas formation of ES2 (k2 and k−2) was less affected. For all affected elementary constants, the same ranking pattern (LF > LF E687D > LF H690A) was observed.
TABLE 4.
Pre-steady-state kinetics of LF15F cleavage by E687D and H690A LF mutants
The pre-steady-state data were obtained by the stopped-flow method. The kinetic parameters were calculated by fitting the experimental data shown in Fig. 3 to Fig. 2B, scheme 2. All kinetic curves were obtained by experiments performed in triplicate and were fitted using Dynafit kinetic software (15).
| Kinetic constants | Substrate type LF15F |
||
|---|---|---|---|
| Enzyme type LF | Enzyme type LF E687D | Enzyme type LF H690A | |
| k1, m−1 × s−1 | (39.1 ± 6.2) × 106 | (12.4 ± 1.1) × 106 | (5.3 ± 1.4) × 106 |
| k−1, s−1 | 245.7 ± 13.5 | 488.4 ± 10.6 | 993.3 ± 244 |
| Kbind1, m−1 | 1.6 × 105 | 2.5 × 104 | 9.6 × 103 |
| k2, s−1 | 3.8 ± 0.1 | 11.7 ± 0.5 | 9.8 ± 1.0 |
| k−2, s−1 | 1.4 ± 0.1 | 9.1 ± 0.2 | 7.7 ± 0.85 |
| Kbind2 | 2.7 | 1.3 | 1.5 |
| k3, s−1 | 0.83 ± 0.01 | 0.18 ± 0.02 | 0.03 ± 0.002 |
| Kp, m | (1.3 ± 0.1) × 10−6 | (9.3 ± 1.2) × 10−6 | (7.3 ± 1.5) × 10−6 |
| Km, m | 2.3 × 10−6 | 17.4 × 10−6 | 42.0 × 10−6 |
| kcat, s−1 | 0.52 | 0.10 | 0.02 |
Cleavage of S20 Peptide Isolated from the Phage Library
Pre-steady-state kinetics was studied for the phage-selected LF substrate S20 (Fig. 1D). The maximum signal acquisition time of the stopped-flow device permits the analysis of S20 binding steps, but cleavage events are beyond the measurable time span (Table 3). Therefore, data for S20 cleavage were collected continuously for 2 h using a conventional spectrofluorimeter (supplemental Fig. 3).
Binding of dansyl-labeled S20 peptide (S20D) followed the same two-step scheme as LF15D (Fig. 2B, scheme 2). The values of the elementary constants calculated for LF15D and S20D interactions with LF suggest that for S20, both binding steps proceed with significantly decreased efficiency compared with LF15 (Table 3). Interestingly, the dissociation rate constants of the first step of the LF-substrate interaction (k−1) were similar for LF15D and S20D. ES1 formation for S20 is ∼10-fold less efficient than for LF15, and ES2 formation is much slower for S20 than for LF15. The ES1-to-ES2 transition proceeds favorably for LF15 (Kbind2 >1), whereas this transition is ineffective for S20 (Kbind2 <1). These observations indicate that discrimination between “good” and “bad” substrates may occur at either step of LF·substrate complex formation.
Activation of the LF Apoenzyme by Divalent Metal Ions
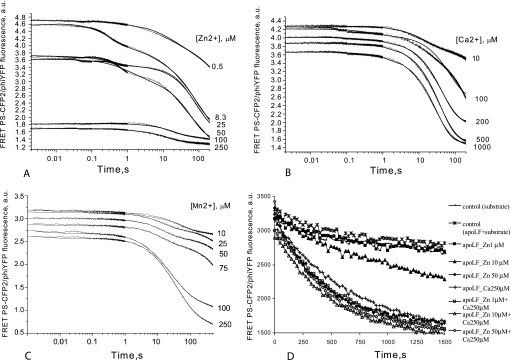
We next examined activation of LF apoenzyme by Zn2+, Ca2+, Mn2+, and Mg2+ in the pre-steady-state mode using the LF15P substrate (Fig. 4). Experimental data obtained during activation of LF apoenzyme with Zn2+ (Fig. 4A) was fitted to the kinetic model shown in (Fig. 2D, scheme 4). The activation curves were biphasic, suggesting that one or two extra Zn2+ ions bind elsewhere within the protein, as was proposed earlier (24). Calcium ions activated apo-LF in a single-step process, with each apo-LF molecule binding a single Ca2+ ion (Fig. 2E, scheme 5). This suggests that calcium can substitute for zinc in the LF active site. Based on the KbindM constants (the sum of the dissociation constants for the elementary steps of metal ion/apo-LF interaction, see Table 5), Zn2+ binding is 325 times more efficient than Ca2+ binding. The rate of substrate hydrolysis is about 1.6 times faster for Zn2+-activated LF compared with Ca2+-activated protease.
FIGURE 4.
Activation of LF apoenzyme by addition of Zn2+ (A), Ca2+ (B), Mn2+ (C), and Zn2+ with Ca2+ (D) was analyzed using LF15P as the substrate. To ensure that the enzyme was inactive at the start of data collection, reaction components (apo-LF and substrate/metal solution) were stored separately in loading syringes and mixed in the measuring cell of the stopped-flow apparatus (for A, B, and C). The concentration of LF15P was 0.25 μm, and the apo-LF concentration was 0.25 μm. Concentrations of divalent metal ions are indicated to the right of the panels. Fig. 2D, scheme 4, was fitted to Zn2+ data, whereas Fig. 2E, scheme 5, describes the mechanisms for Ca2+ and Mn2+. The calculated kinetic parameters are shown in Table 5. D, steady-state kinetics of LF15P cleavage by LF apoenzyme, activated with different concentrations of Zn2+ and Ca2+ and combinations thereof. Legends for respective curves are shown to the right of the graph. The assay was tailored to investigate the possibility of LF activation by low concentrations of Zn2+ in the presence and in the absence of saturating concentration of Ca2+. Concentration of apo-LF was 20 nm, and concentration of LF15P was 1.6 μm. Control reactions contained either uncharged apo-LF with substrate (asterisks) or substrate alone (line marked with small rectangles). Kinetic data for 1, 10, and 100 nm Zn2+ is not shown because of full coincidence with control curves.
TABLE 5.
The pre-steady-state kinetic parameters for substrate hydrolysis by LF apoenzyme activated by divalent metal ions
Pre-steady-state kinetics of substrate cleavage by LF was studied using the stopped-flow method. Prior to reaction initiation, apoenzyme and the mixture containing substrate and divalent metal ions were loaded using two separate syringes to prevent preliminary metal-enzyme and substrate-enzyme interactions. Kinetic parameters were calculated by fitting the experimental data shown in Fig. 4 to Fig. 2D, scheme 4, for Zn2+-activated apoenzyme and Fig. 2E, scheme 5 for Ca2+ and Mn2+ activation. All data were collected in triplicate and fitted using Dynafit software (15). KbindM was calculated as the sum of dissociation constants of each step of metal binding.
| Kinetic constants | Substrate type LF15P |
|||
|---|---|---|---|---|
| Enzyme type LF | Enzyme type apo-LF, metal ion type Zn2+ | Enzyme type apo-LF, metal ion type Ca2+ | Enzyme type apo-LF, metal ion type Mn2+ | |
| k1, m−1 × s−1 | (42.3 ± 6.9) × 106 | (74.6 ± 1.0) × 106 | (88.3 ± 1.1) × 106 | (80 ± 2.0) × 106 |
| k−1, s−1 | 219.3 ± 10.3 | 170.4 ± 2.2 | 270.6 ± 2.7 | 250 ± 5.0 |
| k2, s−1 | 3.9 ± 0.2 | 2.80 ± 0.04 | 2.16 ± 0.03 | 2.0 ± 0.1 |
| k−2, s−1 | 1.3 ± 0.1 | 1.53 ± 0.03 | 1.34 ± 0.02 | 1.1 ± 0.1 |
| k3, s−1 | 0.77 ± 0.04 | 0.71 ± 0.01 | 0.44 ± 0.01 | 0.14 ± 0.01 |
| Km, m | ||||
| Pre-steady state | 1.8 × 10−6 | 1.0 × 10−6 | 1.4 × 10−6 | 1.2 × 10−6 |
| Steady state | (5.4 ± 0.5) × 10−6 | (2.62 ± 0.68) × 10−6 | (3.80 ± 1.28) × 10−6 | (3.67 ± 1.43) × 10−6 |
| kcat, s−1 | ||||
| Pre-steady-state | 0.50 | 0.39 | 0.24 | 0.08 |
| Steady-state | 0.5 ± 0.03 | 0.19 ± 0.02 | 0.18 ± 0.03 | 0.24 ± 0.05 |
| KbindM | 3.9 × 10−6 | 1.2 × 10−4 | 2.2 × 10−5 | |
Although binding of Ca2+ to the LF active site is significantly less efficient than Zn2+ binding, once the protease is saturated by Ca2+, its cleavage efficiency is only slightly inferior to that of Zn2+-charged LF. To study this phenomenon in more detail, we analyzed activation of apo-LF by Mg2+ and Mn2+ in the pre-steady-state mode. Magnesium ions failed to activate apo-LF prepared using extensive dialysis with chelating buffers, whereas divalent manganese ions activated apo-LF as efficiently as Ca2+. Stopped-flow analysis of LF activation by Mn2+ suggests that the activation scheme is similar to that of Ca2+, and fits (Fig. 2E, scheme 5).
The elementary constants presented in Table 5 show that the apo-LF activation ability of divalent ions decreases as follows: Zn2+ > Ca2+ > Mn2+ > Mg2+, with Mg2+ completely unable to activate the enzyme. The binding efficiency of these ions to LF decreases according to the radius of the ion, Zn2+ > Mn2+ > Ca2+.
The fact that the k3 constants for Zn2+, Mn2+, and Ca2+ (the pre-steady-state analogs of stationary kcat) are similar is surprising. Of note, under steady-state conditions, apo-LF charged with Zn2+, Mn2+, and Ca2+ also demonstrated similar kcat values (Table 5).
To completely rule out the possibility that Ca2+ and Mn2+ were contaminated with traces of Zn2+ ions, we collected steady-state kinetic data using apo-LF mixed with Zn2+ at low concentrations, mimicking potential contamination (Fig. 4D). In the range of 1 nm to 1 μm, Zn2+ failed to activate apo-LF. At 10 μm, Zn2+ only partially activates LF, and only 50 μm Zn2+ restores the activity of the apoenzyme to that achieved after incubation with 250 μm Ca2+ (Fig. 4D).
The concentration range in which Zn2+ can activate LF (>1 μm) under steady-state conditions agrees with the KbindM value (3.9 μm) calculated using stopped-flow measurements. Also, the Zn2+ concentrations used in this experiment are well above the levels of Zn2+ contamination in ACS (Mn2+) or molecular biology grade (Ca2+ and Mg2+) reagents used in this study.
DISCUSSION
The function of LF at early stages of anthrax infection is to disable the immune system by disrupting key signal transduction cascades via specific cleavage of MAPKKs (5). Without strict LF substrate specificity, cleavage of off-target substrates would likely lead to activation of the protective mechanisms of the host or to death of the host prior to substantial proliferation of the pathogen. As a result, tight constraints are imposed on the range of substrates cleaved by LF. For example, LF cleaves within the kinase-interacting modules of MAPKKs, but it fails to hydrolyze the structurally and functionally homologous modules in downstream transcription factors such as Jun and Fos (10). Mechanistic insight into the sequence of events during LF-mediated proteolysis will be helpful for understanding the LF substrate discrimination apparatus and for developing assays to identify inhibitors with increased specificity for LF.
Sequences of LF substrates selected from the 8-amino acid randomized substrate phage library were strongly biased toward unprimed (N-terminal to the scissile bond) over primed residues (C-terminal residues), based on alignment with known LF substrates (7). Indeed, the P1′ and sometimes even the P1 residue in phage-selected substrates were encoded by downstream invariant residues of pIII rather than by randomized residues. The most conserved amino acid sequences in phage-selected substrates were RR(V/I), which occupy the same positions (p5, p4, and p3) as the KKV residues in LF15. In the second library iteration, by fixing the conserved substrate positions and by increasing the length of the prime subsite, we isolated the S20 substrate, which was cleaved seven times more efficiently by LF than the MKK6-derived peptide.
The crystal structure of the LF complex with LF15 suggests that basic residues in P4-P5 interact with acidic residues in the binding groove of the protease and that an induced fit mechanism of binding occurs at P1′-S1′ (7). Mutagenesis studies carried out with the S20 substrate suggested that P6, P7, and P2 and P3 of the LF substrate can modulate efficiency of enzyme-substrate interactions, whereas certain combinations of amino acid residues at P1 to P2′ positions are responsible for dramatic changes in kcat. None of S20 mutants harboring single replacement displayed improvement in cleavage efficiency. Moreover, introduction of basic residues at the N terminus of the modified S20 LF substrate can increase efficiency of interaction with LF only if the appropriate context is provided in positions adjacent to the scissile bond (P3 to P1). The position of the scissile bond identified in phage-selected peptide substrates using liquid chromatography/mass spectrometry coincided with the one inferred from structural alignment with known LF substrates.
To further characterize the pattern of LF-substrate interaction suggested by the phage display and mutagenesis studies, we applied a fast kinetics approach that is capable of dissecting the elementary steps of an enzymatic reaction in real time. Elementary constants describing the progression of LF-substrate interactions toward substrate cleavage were calculated based on steady- and pre-steady-state kinetic data collected using LF15 (7) and S20 substrates and three distinct fluorescence-based detection systems (Table 3).
Statistical analysis of LF-substrate interaction data obtained by the stopped-flow technique revealed that LF-mediated proteolysis consists of four elementary steps. Binding of LF to substrate occurred in two steps, ES1 and ES2. Conversion of ES2 to the EP complex (the main chemical stage) represents the rate-limiting reaction step followed by reversible dissociation of reaction products. For all the substrates used, the reaction schemes and elementary constants were similar, further validating the proposed reaction mechanism. Most notably, the two-step binding mechanism was same for S20 and for LF15 substrates. Although it was not possible to detect cleavage of the S20 substrate within the limit of the data collection time using a stopped-flow device, the efficiency of ES1 and ES2 complex formation for S20 was markedly decreased compared with LF15. This suggests that in LF substrate discrimination can occur at both stages of ES formation.
A previous report examined the inhibitory potential of LF15 fragments modified by hydroxamate (7). Only the full LF15 fragment N-terminal to the scissile bond was an efficient inhibitor, whereas shorter fragments immediately adjacent to the scissile were inefficient inhibitors. Addition of positively charged residues to the N terminus of LF peptide substrates can increase binding efficiency, and LF proteolysis is inhibited at high concentration of half-substrates containing nonprime residues (8). Together, these data suggest that initial recognition of the nonprime substrate portion by LF is of critical importance.
Collectively, the above data suggest the following model for LF-substrate interaction. 1) At the first binding step, the KKV or RRV/I, or a similar positively charged motif, binds to LF. Salt bridges between basic and acidic amino acids as well as hydrophobic interactions at P3-S3 mediate the initial substrate recognition. 2) Initial binding may cause conformational changes in LF, leading to exposure of the S1′ pocket. At the second binding step, the hydrophobic amino acid at P1′ inserts into the S1′ pocket, resulting in formation of the productive LF-substrate complex via induced fit (7) and subsequent hydrolysis. Cleavage efficiency strongly depends on the type of adjacent amino acids at P1 and P2′ and positively charged extension (P6 and P7). It is likely that certain combinations of amino acid residues, such as prolines at P1 and P2′, facilitate conformational switch and/or dissociation of leaving groups upon cleavage, whereas a single proline at P1 but not at P2′ may slow down conformational rearrangements in the LF·substrate complex. Data obtained warrants further search for optimal combinations of amino acid residues increasing cleavage susceptibility of LF substrates.
Two-step substrate binding may further increase the selectivity of LF so that only target substrates responsible for specific host defense mechanisms are inactivated without triggering apoptosis or additional host defense systems. Other mechanisms that may contribute to LF cleavage efficiency and specificity, such as exosite interactions, have been described (9). Comparison between the in vivo and in vitro data (10) suggests that exosite interactions can increase the cleavage efficiency of LF by increasing binding of LF to substrate at a remote site, rather than directly affecting the cleavage mechanism.
After detecting a conformational change in LF during ES complex formation, we investigated whether metal ions play a role in this rearrangement. Stopped-flow kinetic analysis of E687D and H690A LF mutants (23) showed that both mutations significantly affected the cleavage rate and the efficiency of ES complex formation. Previous work showed that E687D LF mutant protein is essentially inactive (23), whereas our data indicate that E687D maintains 3% of wild-type LF activity. A plausible explanation for this apparent discrepancy is that the substrate LF15F used in this study was much more susceptible to LF cleavage than earlier peptides, allowing detection of smaller signal changes reflecting residual activity of mutants.
The role of both mutations in decreasing the kcat has been described previously (23), but participation of Zn2+ in ES formation of LF has not. The mechanism of ES stabilization by Zn2+ is not known; either Zn2+ binding affects overall LF conformation and indirectly influences ES1 formation or the metal ion directly interacts with the substrate during the initial binding and conformational rearrangement steps. Crystallographic studies have shown that Zn2+ is likely to stabilize the productive ES complex in thermolysin (25).
Pre-steady-state analysis of LF inhibition by the In-2-LF hydroxamate (20) showed that it binds to LF active sites unoccupied by substrate. Binding of the inhibitor was 2 orders of magnitude more efficient than binding of the best substrate, judging by the Ki versus Km values. Considering the above-discussed possibility of direct interaction between metal ion in the LF active site and a substrate, further investigation of peptide hydroxamates binding to LF, and its mutants can shed light on the role of metal ions at different steps of LF-substrate interaction. Binding of more than one metal ion to a metallopeptidase has been reported previously (26). However, the co-catalytic function of two zinc ions suggested for the Aeromonas aminopeptidase is not supported for LF.
Surprisingly, calcium and manganese ions can also efficiently activate apo-LF, although Ca2+ binds to LF with 325-fold and Mn2+ with 18-fold lower efficiency compared with Zn2+. Despite lower binding affinity of Ca2+ and Mn2+ to LF, apo-LF saturated with either of these ions cleaves LF15P substrate with almost the same efficiency as Zn2+-charged LF. Titration of apo-LF by Zn2+ ruled out the possibility that the observed activation is because of Zn2+ contamination of the Ca2+ and Mn2+ salts used in this study. Furthermore, Mg2+ in the same concentration range completely failed to activate apo-LF. Although the extracellular calcium concentration in humans is 1.0–1.2 mm, intracellular calcium concentration is 1000 times lower even after calcium release during certain types of cell activation (27). The mechanism and physiological relevance of LF activation by Ca2+ require further study.
Here we analyzed the mechanism of LF-substrate interaction using combinatorial and kinetic approaches. By analyzing pre-steady-state kinetics, we identified the elementary steps of the proteolytic reaction catalyzed by LF. A two-step binding mechanism similar to the one that brings the LF·substrate complex into a substrate proteolysis conformation has been proposed earlier for thermolysin (28) and human immunodeficiency virus, type 1, protease (29).
The finding that binding of LF to substrate occurs in at least two steps to yield a productive pre-cleavage complex may facilitate the development of efficient LF inhibitors. Selectivity is one of the most important features of classic metalloprotease inhibitors. An inhibitor capable of freezing the transition of LF to the productive conformation would have a significant advantage over classic inhibitors in terms of specificity. Such inhibitors that immobilize an enzyme in a “dead” conformation have been recently described for protein kinases, and they offer greater target specificity compared with molecules targeting the ATP-binding site (30). For instance, imatinib, one of the most successful protein kinase inhibitors, causes the Abelson tyrosine kinase to adopt an inactive conformation (31). Application of the stopped-flow approach helped us to dissect the mechanistic details of hydroxamate-based inhibition. Likewise, analysis of the pre-steady-state kinetics of LF-mediated proteolysis in the presence of small molecules targeting LF can help to identify and further optimize LF inhibitors that offer greater selectivity and inhibitory capacity compared with conventional metalloprotease inhibitors.
Supplementary Material
Acknowledgments
We thank Dr. Eric A. E. Garber (Food and Drug Administration, Center for Food Safety and Applied Nutrition, College Park, MD), Dr. Carlton D. Jackson (National Center for Toxicological Research, Jefferson, AR), and Dr. Paul Wentworth, Jr. (The Scripps Research Institute, La Jolla, CA), for helpful discussions and critical reading of the manuscript. We also thank Dr. Rustam Kh. Ziganshin for carrying out mass spectrometry experiments.
This work was supported by United States Civilian Research & Development Foundation Grant 2496, NATO Grant SfP 980870, and the Russian Ministry of Science Grants 02.512.11.2007 and 02.512.11.2208.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I and II and Figs. 1–4.
- PA
- protective antigen
- LF
- lethal factor
- FRET
- fluorescent resonance energy transfer
- ELISA
- enzyme-linked immunosorbent assay
- HPLC
- high pressure liquid chromatography
- mAb
- monoclonal antibody
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl
- MAPKK
- mitogen-activated protein kinase kinase.
REFERENCES
- 1.Allerberger F., Grif K., Dierich M. P., Wimmer A., Plicka H. ( 2002) Lancet 359, 711. [DOI] [PubMed] [Google Scholar]
- 2.Jernigan D. B., Raghunathan P. L., Bell B. P., Brechner R., Bresnitz E. A., Butler J. C., Cetron M., Cohen M., Doyle T., Fischer M., Greene C., Griffith K. S., Guarner J., Hadler J. L., Hayslett J. A., Meyer R., Petersen L. R., Phillips M., Pinner R., Popovic T., Quinn C. P., Reefhuis J., Reissman D., Rosenstein N., Schuchat A., Shieh W. J., Siegal L., Swerdlow D. L., Tenover F. C., Traeger M., Ward J. W., Weisfuse I., Wiersma S., Yeskey K., Zaki S., Ashford D. A., Perkins B. A., Ostroff S., Hughes J., Fleming D., Koplan J. P., Gerberding J. L. ( 2002) Emerg. Infect. Dis. 8, 1019– 1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mock M., Fouet A. ( 2001) Annu. Rev. Microbiol. 55, 647– 671 [DOI] [PubMed] [Google Scholar]
- 4.Young J. A., Collier R. J. ( 2007) Annu. Rev. Biochem. 76, 243– 265 [DOI] [PubMed] [Google Scholar]
- 5.Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., Vande Woude G. F. ( 1998) Science 280, 734– 737 [DOI] [PubMed] [Google Scholar]
- 6.Pannifer A. D., Wong T. Y., Schwarzenbacher R., Renatus M., Petosa C., Bienkowska J., Lacy D. B., Collier R. J., Park S., Leppla S. H., Hanna P., Liddington R. C. ( 2001) Nature 414, 229– 233 [DOI] [PubMed] [Google Scholar]
- 7.Turk B. E., Wong T. Y., Schwarzenbacher R., Jarrell E. T., Leppla S. H., Collier R. J., Liddington R. C., Cantley L. C. ( 2004) Nat. Struct. Mol. Biol. 11, 60– 66 [DOI] [PubMed] [Google Scholar]
- 8.Tonello F., Ascenzi P., Montecucco C. ( 2003) J. Biol. Chem. 278, 40075– 40078 [DOI] [PubMed] [Google Scholar]
- 9.Chopra A. P., Boone S. A., Liang X., Duesbery N. S. ( 2003) J. Biol. Chem. 278, 9402– 9406 [DOI] [PubMed] [Google Scholar]
- 10.Liang X., Young J. J., Boone S. A., Waugh D. S., Duesbery N. S. ( 2004) J. Biol. Chem. 279, 52473– 52478 [DOI] [PubMed] [Google Scholar]
- 11.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. ( 1989) Gene 77, 51– 59 [DOI] [PubMed] [Google Scholar]
- 12.Novagen ( 1997) pET System Manual, 7th Ed, Vol. 4, p. 49, Novagen, Madison, WI [Google Scholar]
- 13.Souslova E. A., Chudakov D. M. ( 2006) Microsc. Res. Tech. 69, 207– 209 [DOI] [PubMed] [Google Scholar]
- 14.Cummings R. T., Salowe S. P., Cunningham B. R., Wiltsie J., Park Y. W., Sonatore L. M., Wisniewski D., Douglas C. M., Hermes J. D., Scolnick E. M. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 6603– 6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzmic P. ( 1996) Anal. Biochem. 237, 260– 273 [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsov N. A., Koval V. V., Nevinsky G. A., Douglas K. T., Zharkov D. O., Fedorova O. S. ( 2007) J. Biol. Chem. 282, 1029– 1038 [DOI] [PubMed] [Google Scholar]
- 17.Kuznetsov N. A., Koval V. V., Zharkov D. O., Vorobjev Y. N., Nevinsky G. A., Douglas K. T., Fedorova O. S. ( 2007) Biochemistry 46, 424– 435 [DOI] [PubMed] [Google Scholar]
- 18.Vitale G., Bernardi L., Napolitani G., Mock M., Montecucco C. ( 2000) Biochem. J. 352, 739– 745 [PMC free article] [PubMed] [Google Scholar]
- 19.Smith M. M., Shi L., Navre M. ( 1995) J. Biol. Chem. 270, 6440– 6449 [DOI] [PubMed] [Google Scholar]
- 20.Tonello F., Seveso M., Marin O., Mock M., Montecucco C. ( 2002) Nature 418, 386. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K., Yaoita Y. ( 1997) Nucleic Acids Res. 25, 2231– 2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobb R. R., Auld D. S. ( 1980) Biochemistry 19, 5297– 5302 [DOI] [PubMed] [Google Scholar]
- 23.Hammond S. E., Hanna P. C. ( 1998) Infect. Immun. 66, 2374– 2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kochi S. K., Schiavo G., Mock M., Montecucco C. ( 1994) FEMS Microbiol. Lett. 124, 343– 348 [DOI] [PubMed] [Google Scholar]
- 25.Holland D. R., Hausrath A. C., Juers D., Matthews B. W. ( 1995) Protein Sci. 4, 1955– 1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevrier B., Schalk C., D'Orchymont H., Rondeau J. M., Moras D., Tarnus C. ( 1994) Structure 2, 283– 291 [DOI] [PubMed] [Google Scholar]
- 27.Nussey S. S., Whitehead S. A. ( 2001) Endocrinology. An Integrated Approach, pp. 173– 174, BIOS Scientific Publishers Ltd., Oxford, UK: [PubMed] [Google Scholar]
- 28.Mock W. L., Stanford D. J. ( 1996) Biochemistry 35, 7369– 7377 [DOI] [PubMed] [Google Scholar]
- 29.Scott W. R., Schiffer C. A. ( 2000) Structure 8, 1259– 1265 [DOI] [PubMed] [Google Scholar]
- 30.Rellos P., Ivins F. J., Baxter J. E., Pike A., Nott T. J., Parkinson D. M., Das S., Howell S., Fedorov O., Shen Q. Y., Fry A. M., Knapp S., Smerdon S. J. ( 2007) J. Biol. Chem. 282, 6833– 6842 [DOI] [PubMed] [Google Scholar]
- 31.Schindler T., Bornmann W., Pellicena P., Miller W. T., Clarkson B., Kuriyan J. ( 2000) Science 289, 1938– 1942 [DOI] [PubMed] [Google Scholar]
- 32.Huang C. Y. ( 1979) Methods Enzymol. 63, 54– 84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.