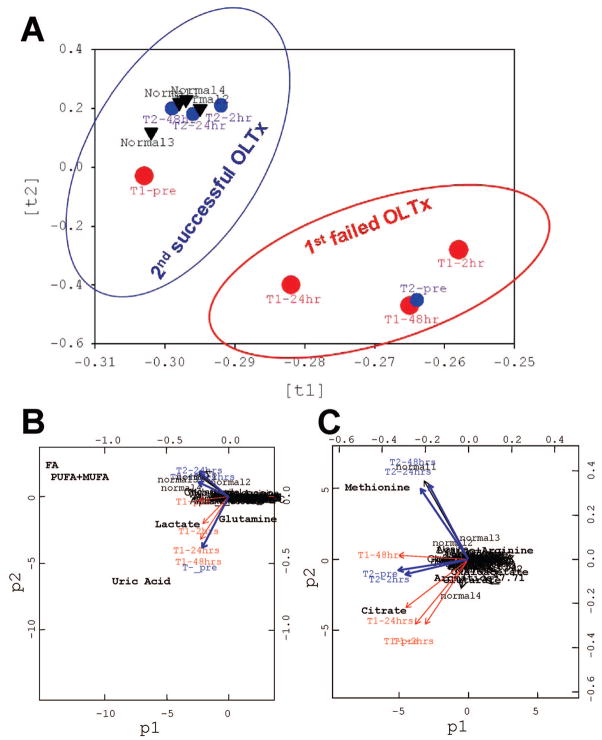
FIGURE 1.
Principle component analysis (PCA): (A) PCA scores (ti) on global metabolic pattern to visualize group clustering between the first failed and second successful OLTx compared to the normal blood profile; (B) PCA plots (pi) on absolute metabolite concentrations and (C) on normalized metabolite concentrations to distinguish single putative biomarkers responsible for cloistering pattern on the loading plot in (A). In the first step, the PCA scores (ti,) described the variation in the sample direction, i.e. similarity/dissimilarity between samples. This step allowed for pattern recognition (normal/abnormal group assignment in A). Each point (in circles or triangles) represent a metabolic profile from each samples. In the next step, the PCA loading plots (pi) described the variation in the variable direction, i.e. similarity/dissimilarity between variables. This step explained the patterns in previous scores (ti) and identified metabolites which were responsible for the group clustering (B, C). The metabolites condensed in the center do not contribute to group clustering in (A). The “outliner” metabolites differ among the group. All mathematical models were built with R package (2.00). First OLTx (graft failure, big red circles) time points: T1-pretransplant; T1-2hr after first transplantation; T1-24hr; and T1-48-hr; and second OLTx (functional graft, blue small circles) time points: T2-pre; T2-2hr after second transplantation; T2-24hr; and T2-48hr. Four blood profiles from healthy volunteers (normal 1, 2, 3, and 4, black triangles) are clustering with the data points from the second transplant. All data are based on PCA of blood metabolic profiles.