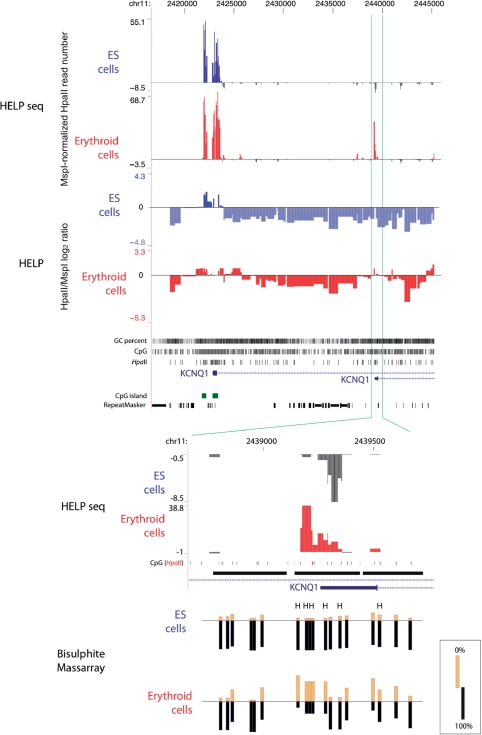
Figure 6.
Adaptation of the HELP assay to use massively parallel sequencing: HELP-seq. Instead of using microarrays to map the representations to their genomic source, we sequenced the ends of the fragments using massively parallel sequencing (Illumina). We show the promoter region of the KCNQ1 gene as an example of the concordant data obtained, with hypomethylated sites represented as an upward/positive peak and methylated as downward/negative, in both the microarray-based (HELP) and Illumina-based (HELP-seq) assays. The HELP data are represented by quantile-normalized HpaII/MspI log2 ratios (13) while the HELP-seq data are represented by the number of HpaII reads per locus normalised to MspI reads for the same locus as described in the main text. Both embryonic stem (ES, red) and derived in vitro-differentiated erythroid progenitor (blue) cells are shown. The upstream (left) promoter of KCNQ1 is hypomethylated in both cell types, with a broader region of hypomethylation in erythroid compared with ES cells in both the HELP and HELP-seq studies. The downstream promoter appears to be methylated in ES cells in both assays, but microarray-based HELP weakly indicates hypomethylation in erythroid cells, whereas HELP-seq shows a strong hypomethylated signal in these cells. To determine which technique was more accurate, we performed validation using bisulphite MassArray (bottom), confirming the underlying hypomethylation of this locus in the erythroid and not ES cells, and demonstrating that HELP-seq offers improved capacity to identify sites of hypomethylated DNA in the genome.