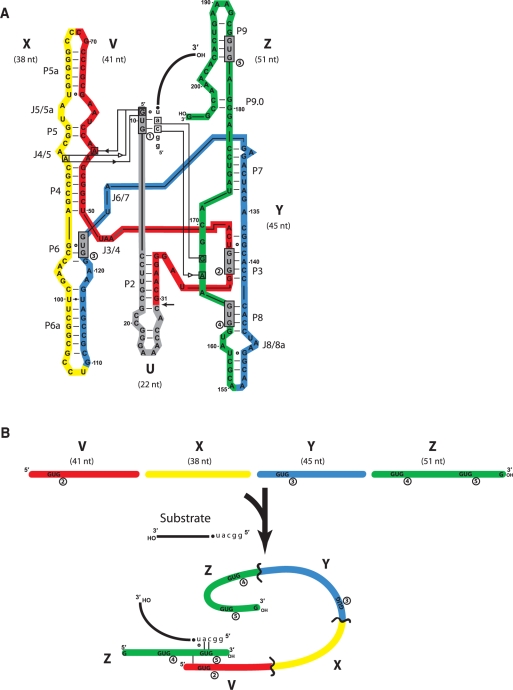
Figure 1.
Schematic of the partitioning of the Azoarcus ribozyme into fragments. (A) The 197-nt source molecule was partitioned into five fragments (U, gray; V, red; X; yellow; Y, blue; Z, green) such that catalytic activity in trans, plus covalent self-assembly could be assayed. Removal of the U fragment from the system at the location indicated by the arrow leaves an L–30 construct which can then be divided into four fragments that can exhibit activity when 5-nt head groups (h = GGCAU) are appended to the 5′ portions of X, Y and Z (see text). A substrate oligomer (lower-case letters = head; black line = tail) binds via a 5′-CAU-3′ to the IGS (GUG) of the ribozyme, which catalyzes phosphotransfer of the tail to its own 3′ end. The native IGS of the ribozyme is denoted by GUG1, while the four exogenous GUG triplets that occur naturally in the remainder of the ribozyme are gray boxes denoted 2–5 (in circles). The tertiary interactions that hold the IGS and the IGS complement into the active site as determined by X-ray crystallography (24) are denoted using the hydrogen-bonding symbolism of Leontis et al. (41). (B) Schematic of how a ribozyme complex (either as a covalently contiguous molecule or as several fragments cooperating in trans) can perform catalysis in the absence of the U fragment that contains the native IGS. Here, the use of an exogenous IGS (GUG5) present in the h•Z fragment to bind to the IGS complement CAU is depicted.