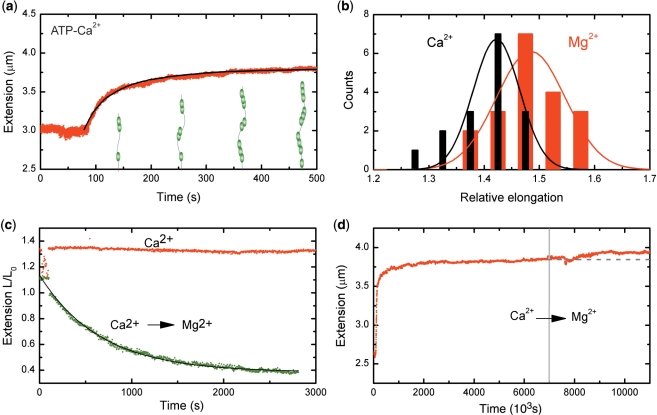
Figure 2.
RecA–ssDNA filaments in conditions suppressing ATP hydrolysis. (a) Filaments formed on a 8.6 kb ssDNA molecule showed a similar binding behavior at a stretching force of 6 pN in a 5 mM Ca2+ buffer to that in the presence of Mg2+. The black line is a fit from MC simulations, adopting hexamers for nucleation and extension units, used to determine the nucleation and extension rate. (b) Histogram of RecA–ssDNA tether length at a stretching force of 6 pN in the presence of either 5 mM Mg2+ (red) or 5 mM Ca2+ (black). Solid lines are a fit to a normal distribution. (c) RecA disassembly at a stretching force of 0.5 pN from a 8.6 kb ssDNA was blocked in the presence of 5 mM Ca2+ (red points), but readily occurred when Ca2+ was replaced by Mg2+ (green points) The filament dissociation fits well to a single exponential decay (black line). (d) A 7.3 kb RecA–ssDNA filament assembled in the presence of Ca2+ at a stretching force of 6 pN showed a further increase in end-to-end distance when the buffer was exchanged for one containing 1 µM RecA and 5 mM Mg2+. The gray dashed line is a guide to the eye.