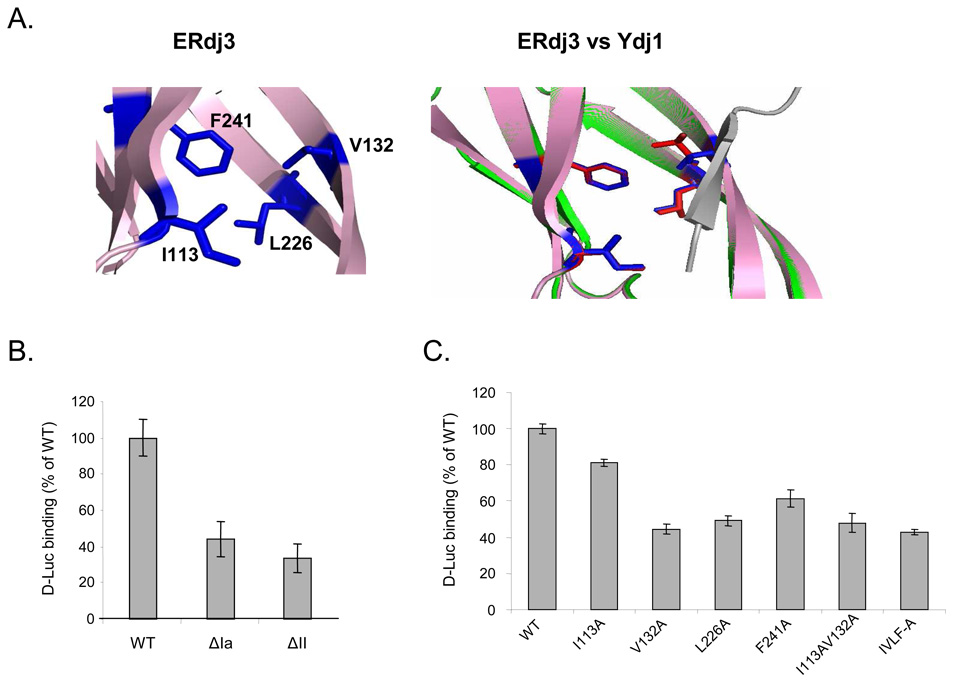
Figure 3. Both domain I and domain II of ERdj3 contribute to its ability to bind to denatured luciferase.
(A) Model of ERdj3’s putative substrate binding domain (left) shows amino acid residues that could form a structure similar to that found on Ydj1. An overlap of the peptide binding site of Ydj1 with the model of the corresponding region of ERdj3 is shown on right. Grey: substrate peptide, green: Ydj1, pink: ERdj3 model, red: peptide interacting residues on Ydj1, and blue: corresponding residues on ERdj3. (B) Measurement of complex formation between wild-type ERdj3 and the ΔIa and ΔII +GSGG mutants and denatured luciferase was performed as described in the Materials and Methods section. The quantity of luciferase bound to wild-type ERdj3 was set to 100%, and the values for the various mutants were expressed as a percent of this value. Standard errors are indicated. (C) Measurement of complex formation between the indicated ERdj3 point mutants and luciferase.