Figure 5.
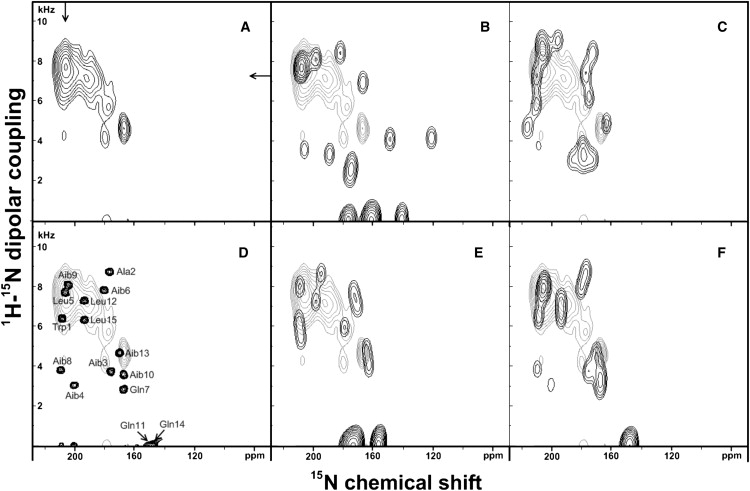
Experimental and simulated PISEMA spectra for 15N uniformly labeled ampullosporin A reconstituted into oriented di-C12:0-PC at a peptide/lipid ratio of 1/75. (A) Experimental spectrum. (B) Simulated spectrum of the x-ray diffraction structure of ampullosporin A (pdb: AMPA) superimposed on the experimental spectrum (light gray); (C) Simulation of an ideal α-helix (ϕ = −65°, ψ = −45°). (D) The simulation shown in panel F is represented with reduced line-broadening to resolve the peaks and to monitor the origin of the simulated signal intensities. (E) Simulated spectrum of an ideal 310-helix (ϕ = −50°, ψ = −31°). (F) simulation of a mixed α-/310-helical peptide (the first eight residues folded in α-helical and the last seven residues in 310-helical conformation). The simulation details are summarized in Table 2.
