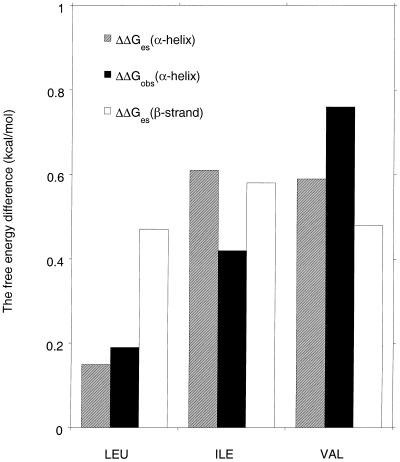
Figure 4.
Comparison between the measured helix propensity difference (6) (filled bars) and the calculated ESF difference for a mutant peptide versus an alanine host in either the β-strand conformation (empty bars) or the helical conformation (hatched bars). The units are kcal/mol. The comparison is made for Leu, Ile, and Val. The β-strand peptides contain five alanine residues plus N-terminal acetyl and C-terminal N-methylamide blocking groups, and only the central Ala residue is substituted by another amino acid. The ESF values of the five central residues of the mutant β-strand peptide are subtracted from those of the Ala peptide and then summed. The peptide sequence used for calculating ESF values in the helical conformation corresponds to the peptide sequence used for measuring helix propensities (6), which is Ac-K(A)4X(A)4KGY-NH2, where X is either A, L, I, V, or G. In calculating ESF differences between helical peptides, the values for the five central peptide groups (groups 4–8) in the mutant peptide are subtracted from the corresponding values for the Ala peptide and then summed. The side-chain torsion angles are: Leu, χ1–60, χ2 180°; Ile, χ1–60, χ2 180°; Val, χ1 180°; the backbone angles are given in the legends to Figs. 1 and 2.