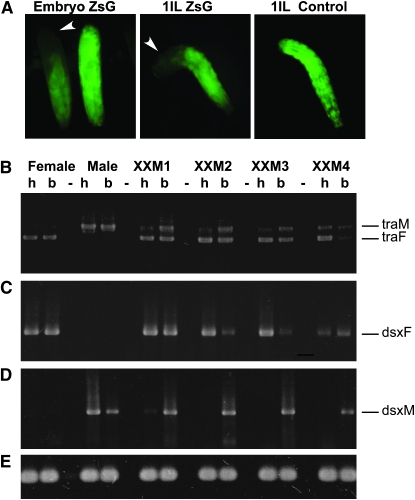
Figure 5.—
Analysis of the splicing patterns of Lctra and Lcdsx in XX transformed males and wild-type individuals. (A) Control for the RNAi technique; embryos from a cross between Lchsp83-ZsGreen males and wild-type females were injected with dsRNA for ZsGreen. Fluorescence is lost in the posterior end of injected embryos and larvae but not in the anterior end. (B) RT–PCR amplification with Lctra-specific primers on total RNA isolated from heads (h) and bodies (thorax plus abdomen) (b) of transformed XX males presenting male genitalia and female head (XXM1, XXM2, and XXM3), a completely transformed XX male (XXM4) and of a wild-type female and male. The primers used amplify different size products for male (traM) and female transcripts (traF). (C and D) RT–PCR amplification with Lcdsx female and male specific primers, using the same total RNA samples as in B. The sex-specific amplification products are labeled dsxF for female and dsxM for male, respectively. (E) RT–PCR control using α-tubulin-specific primers. The RNAi knockdown of Lctra in XX males sets Lctra splicing in the male mode in the posterior end of injected individuals, which in turn changes the splicing pattern of Lcdsx from the female to the male form.