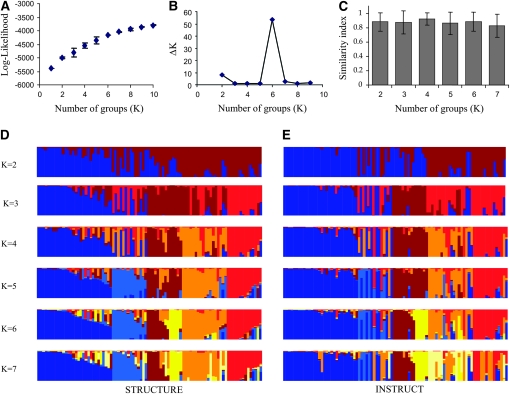
Figure 1.—
Analysis of the population structure of pearl millet inbred lines. The analysis of population structure in inbred lines was performed using STRUCTURE (Pritchard et al. 2000b; Falush et al. 2003) and INSTRUCT (Gao et al. 2007). (A) The average log-likelihood and the standard error of 10 different runs of STRUCTURE were calculated. The log-likelihood showed a steady increase as the number of groups (K) increased, and no clear maxima were detected. (B) To assess the number of groups (K) supported by the analysis, we also calculated the second-order change in the log-likelihood ΔK (Evanno et al. 2005). A clear change was detected for K = 6, suggesting six was the number of groups supported by the STRUCTURE analysis. To allow a comparison of the two Bayesian structure inference methods of STRUCTURE and INSTRUCT, we calculated a similarity index (see text for details). (C) The average similarity index and standard error values for each individual were reported. The average similarity index was >80% in most cases, suggesting similar inference of ancestry results for each plant. We finally represented the results of a run of STRUCTURE (D) and INSTRUCT (E) to enable direct visual comparison of the two methods. The graph (D) represents the run with the highest likelihood of STRUCTURE for a number of populations (K), and E represents the run of INSTRUCT that showed the lowest deviance information criterion (DIC). The ancestry (q) of each of the inbred lines in a population is represented by a different color. The different colors correspond to the different populations identified by STRUCTURE and INSTRUCT. The global visual comparison highlighted a global similarity, but some differences were clearly observed between the two analyses.