Abstract
Idiopathic hypogonadotropic hypogonadism (IHH) is a condition characterized by failure to undergo puberty in the setting of low sex steroids and low gonadotropins. IHH is due to abnormal secretion or action of the master reproductive hormone gonadotropin-releasing hormone (GnRH). Several genes have been found to be mutated in patients with IHH, yet to date no mutations have been identified in the most obvious candidate gene, GNRH1 itself, which encodes the preprohormone that is ultimately processed to produce GnRH. We screened DNA from 310 patients with normosmic IHH (nIHH) and 192 healthy control subjects for sequence changes in GNRH1. In 1 patient with severe congenital nIHH (with micropenis, bilateral cryptorchidism, and absent puberty), a homozygous frameshift mutation that is predicted to disrupt the 3 C-terminal amino acids of the GnRH decapeptide and to produce a premature stop codon was identified. Heterozygous variants not seen in controls were identified in 4 patients with nIHH: 1 nonsynonymous missense mutation in the eighth amino acid of the GnRH decapeptide, 1 nonsense mutation that causes premature termination within the GnRH-associated peptide (GAP), which lies C-terminal to the GnRH decapeptide within the GnRH precursor, and 2 sequence variants that cause nonsynonymous amino-acid substitutions in the signal peptide and in GnRH-associated peptide. Our results establish mutations in GNRH1 as a genetic cause of nIHH.
Keywords: GnRH, luteinizing hormone-releasing hormone, LHRH
Gonadotropin-releasing hormone (GnRH) is the master hormone of the reproductive endocrine system. The existence of central hormones that regulate reproduction was postulated a century ago (reviewed in ref. 1). In 1910, Crowe et al. (2) demonstrated that disruption of the hypothalamic-pituitary connection in dogs prevented the onset of puberty. Subsequent studies led to the hypothesis that the pituitary is controlled by a hypothalamic factor (3–6). It was not until 1971, however, that the amino acid sequence of GnRH was determined after extraction from the hypothalami of thousands of pigs and sheep by the groups of Schally and Guillemin (7, 8).
The neurons that secrete GnRH arise in the olfactory placode and migrate into the hypothalamus (reviewed in ref. 1). Once in the hypothalamus, these GnRH neurons project axons to the median eminence and synchronize their secretion of GnRH in a pulsatile fashion. GnRH is then carried by the portal circulation to the pituitary gland and stimulates the gonadotropes of the anterior pituitary to secrete the gonadotropins follicle-stimulating hormone (FSH) and luteinizing hormone (LH). These gonadotropins then evoke steroidogenesis and gametogenesis from the gonads.
Complementing extensive physiologic studies of the central role of GnRH in reproduction (reviewed in ref. 1), human genetic studies have underscored the critical role of GnRH in regulating reproduction (reviewed in refs. 9, 10). Idiopathic hypogonadotropic hypogonadism (IHH) is characterized by the absence of spontaneous pubertal development in the face of low sex steroid and gonadotropin levels with otherwise normal pituitary function. When associated with anosmia, this hypogonadotropism is termed Kallmann syndrome (KS), whereas isolated hypogonadotropic hypogonadism with a normal sense of smell is termed normosmic IHH (nIHH). Studies of patients with nIHH and KS have led to the identification of several genes that regulate reproduction. Mutations in KAL1 (11, 12), FGFR1 (13), FGF8 (14), PROK2 (15), PROKR2 (15), and CHD7 (16) are thought to disrupt the development and migration of GnRH neurons, thereby resulting in KS and/or nIHH. Patients with mutations in PCSK1, which encodes prohormone convertase 1/3, exhibit hypogonadotropic hypogonadism because of abnormal processing of the GnRH decapeptide from its prohormone precursor (17). Mutations in GPR54 cause nIHH by interfering with the normal secretion of GnRH (18, 19), and mutations in GNRHR, which encodes the GnRH receptor, result in inability to respond to GnRH (20). Mutations in the genes TAC3 and TACR3, which encode neurokinin B and its receptor, respectively, have recently been implicated in nIHH (21), although their precise functions in reproduction remain unclear.
A glaring omission from the list of genes implicated in IHH is GNRH1 itself, which encodes the preprohormone that is ultimately processed to produce GnRH. Findings in the mouse certainly suggest that human mutations in GNRH1 would cause nIHH. The hpg mouse carries a deletion of Gnrh1 that arose spontaneously and results in complete absence of GnRH synthesis (22, 23). Male and female hpg mice are sexually infantile, infertile, and exhibit low sex steroid and gonadotropin levels (22). In one of the earliest demonstrations of successful gene therapy, the reproductive deficits of hpg mice were rescued by a Gnrh1 transgene (24). Aside from their reproductive phenotypes, hpg mice appear grossly normal, although dental abnormalities have recently been reported (25). The clear association of loss of Gnrh1 function in the mouse with hypogonadotropic hypogonadism makes the absence of human GNRH1 mutations as a cause of nIHH all the more puzzling.
We herein report a homozygous mutation in a male patient with severe congenital nIHH. This single base-pair deletion produces a frameshift that is predicted to disrupt the GnRH decapeptide. We also identified rare heterozygous GNRH1 sequence variants in 4 patients with nIHH.
Results
Patient Phenotypes.
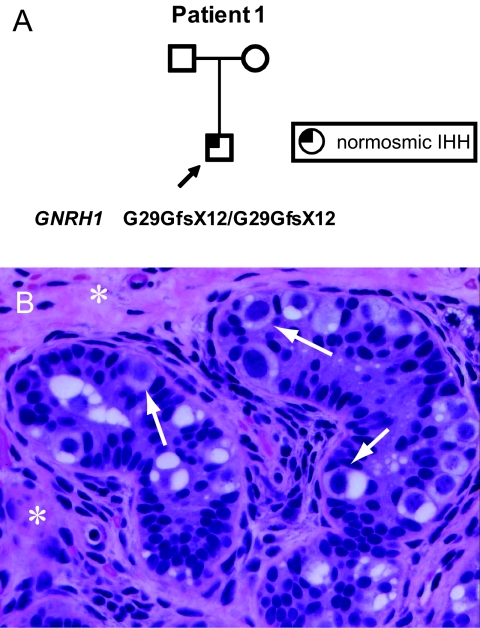
Patient 1.
Patient 1 was evaluated at 8 years, 8 months, for cryptorchidism and microphallus. His parents had normal pubertal timing, come from the same village in Armenia, and deny consanguinity (Fig. 1A). The patient's examination was notable for height between the 10th and 25th percentiles, weight between the 75th and 90th percentiles, nonpalpable testes, microphallus (<3 cm), and absence of midline or skeletal defects. FSH and LH were both <0.5 international units (IU)/L, and anti-Müllerian hormone was 99 pmol/L (reference range for prepubertal boys, 100–300 pmol/L), indicating the presence of testicular Sertoli cells. A human chorionic gonadotropin (hCG) stimulation test at 9 years produced no change in serum testosterone (from 0.38 to 0.4 nmol/L). Ultrasound identified inguinal testes with calculated volumes of 0.13 and 0.11 mL. Shortly thereafter, he underwent bilateral orchiopexy. Intraoperative bilateral testicular biopsies showed immature seminiferous tubules with no lumen, gonocyte-like cells, immature Sertoli cells, and interstitial fibrosis with spindle-shaped myofibroblasts (Fig. 1B).
Fig. 1.
Features of patient 1, who has a homozygous frameshift mutation in GNRH1. (A) Pedigree. Arrow, proband. (B) Testicular biopsy of patient 1, stained with hematoxylin and eosin. [Magnification: 1,000×.] Arrows, gonocyte-like cells; asterisks, interstitial cells.
At age 13 years, 6 months, a GnRH stimulation test was performed because of lack of pubertal development. Baseline FSH and LH were both <0.5 IU/L and rose minimally to 1.3 and 0.8 IU/L, respectively. Formal smell testing with a set of odorants revealed a normal sense of smell. He began treatment with testosterone enanthate 100 mg every 4 weeks, which resulted in linear growth and development of secondary sexual characteristics. Currently, at age 15 years, 6 months, his height is 168 cm (50th percentile), his weight is 81 kg, and he has developed facial hair and Tanner V pubic hair.
Patient 2.
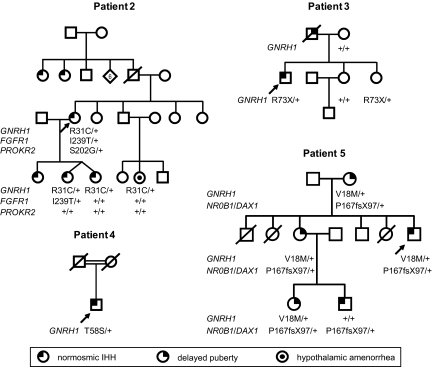
Patient 2 presented at 19 years with absence of breast development and was diagnosed with nIHH. She conceived 1 singleton and 1 twin pregnancy after gonadotropin treatment (Fig. 2). At age 42 years, she exhibited reversal of IHH, with normal menstrual cycles after stopping hormonal therapy.
Fig. 2.
Pedigrees of patients with nIHH found to have heterozygous sequence variants in GNRH1. Arrows, probands; +, wild type.
The patient's eldest daughter presented at 14 years with primary amenorrhea and Tanner III breast development, whereas her younger twin daughters were started on estrogen therapy at 13 years for absent puberty (Fig. 2). The diagnosis of nIHH was confirmed in her daughters when they failed to resume menses after discontinuation of hormonal therapy at age 18. Patient 2's two paternal aunts also have nIHH, and her niece has hypothalamic amenorrhea (Fig. 2).
Genetic analysis of this patient identified a heterozygous mutation in FGFR1 (p.I239T) and a heterozygous variant in PROKR2 (p.S202G) (Table 1 and Fig. 2). The patient's twin daughters and niece do not carry the PROKR2 variant, and one of the patient's daughters carries the FGFR1 mutation (Fig. 2).
Table 1.
Phenotypes of nIHH probands with GNRH1 variants
| Subject | Sex | Base pair change | Amino acid change | Ethnicity | Inheritance | Notable phenotypes | Other gene variants | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1898001 | M | [c.87delA] + [c.87delA] | [p.G29GfsX12] + [p.G29GfsX12] | Caucasian | Sporadic | Bilateral cryptorchidism, microphallus | None identified |
| 2 | 79701 | F | [c.91C > T] + [=] | [p.R31C] + [=] | Caucasian | Familial | Reversal at age 42 | FGFR1, PROKR2 |
| 3 | 08901 | M | [c.217C > T] + [=] | [p.R73X] + [=] | Caucasian | Familial | Right retractile testis, microphallus | None identified |
| 4 | 1820001 | M | [c.172A > T] + [=] | [p.T58S] + [=] | Asian | Sporadic | Microphallus, retinitis pigmentosa | None identified |
| 5 | 12301 | M | [c.52G > A] + [=] | [p.V18M] + [=] | Caucasian | Familial | Adrenal insufficiency | NR0B1/DAX1 |
F, female; M, male.
Patient 3.
Patient 3 was a product of a normal delivery at term after a benign pregnancy. He had descended testes at birth, but the left testis was retractile at 5 days. At 7 years, he underwent left inguinal hernia repair and left orchiopexy. Acne developed at age 11. He had a history of febrile seizures and received phenobarbital prophylaxis until age 14. His father was reported to be a “delayed bloomer” (Fig. 2).
The patient was evaluated at 17 years, 6 months, for absent puberty. His physical examination was notable for a high-pitched voice, mild acne on the chin, absence of facial hair, sparse axillary hair, a few strands of pubic hair, microphallus (length 2.5 cm), right testicle with length of 2 cm and soft consistency, left testicle with length of 1 cm, and intact olfaction as tested by serial dilutions of pyridine solutions. Laboratory evaluation was notable for testosterone, 1.9 nmol/L; FSH, 2 IU/L; and LH, 4–6 IU/L. Karyotype was 46,XY. After an hCG stimulation test (5,000 IU daily for 5 days), testosterone rose from 1.9 to 3.0 nmol/L. After a clomiphene stimulation test, FSH, LH, and testosterone were essentially unchanged. Bone age was 15 years. He was treated with escalating doses of intramuscular testosterone for 10 months and reported little response.
At age 22 years, 5 months, the patient underwent detailed laboratory evaluation at Massachusetts General Hospital (MGH). Aside from hypogonadotropic hypogonadism, he had normal anterior pituitary function. Frequent sampling for 12 h showed absence of LH pulses (Fig. S1A). A 7-day challenge with pulsatile GnRH resulted in progressive increases in FSH and LH, but his testosterone level remained low (Fig. S1B). A computed tomography scan of the sella was normal. A right testicular biopsy performed at age 22 years, 11 months, showed small seminiferous tubules and complete absence of Leydig cells, consistent with gonadotropin deficiency. Bone age was 17 years. Subsequent treatment with pulsatile GnRH therapy for >1 year resulted in normalization of serum testosterone levels, elevated gonadotropin levels, and increase in testicular size to 5–6 mL bilaterally.
Patient 4.
Patient 4 was born to Chinese/Cambodian parents who were first cousins (Fig. 2), and he had a small penis at birth. He was evaluated at 16 years for absence of secondary sexual characteristics. Treatment with escalating doses of intramuscular testosterone enanthate induced development of secondary sexual characteristics. At 26 years, testes were 3 mL bilaterally. Smell testing with the University of Pennsylvania Smell Identification Test revealed a normal sense of smell. The patient also has progressive visual loss that started at 9 years because of retinitis pigmentosa.
Patient 5.
Patient 5's detailed medical and family history has been reported in ref. 26. The patient and his nephew are hemizygous for a frameshift mutation in NR0B1 (also called DAX1) (Table 1 and Fig. 2) and consequently have X-linked adrenal hypoplasia congenita, with primary adrenal insufficiency and nIHH. The patient's mother, sister, and niece had delayed puberty and are heterozygous for the NR0B1/DAX1 mutation (Fig. 2 and Table 1).
Sequence Changes in GNRH1.
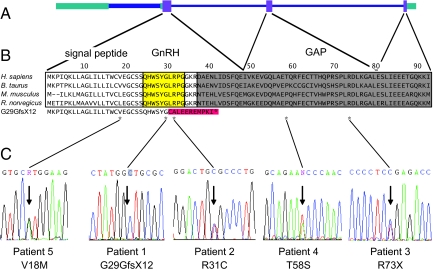
GNRH1 consists of 3 coding exons (Fig. 3), which encode a 92-aa preprohormone that is processed to produce the GnRH decapeptide and the GnRH-associated peptide (GAP) (1). DNA from 310 patients with nIHH and 192 control subjects underwent sequencing of the GNRH1 exons and exon–intron boundaries.
Fig. 3.
Sequence variants in GNRH1 identified in patients with nIHH. (A) Genomic structure of GNRH1. Purple boxes, coding regions of exons; green boxes, noncoding regions of exons; narrow blue lines, introns; thick blue line, a variant intron within the first exon. (B) Alignment of amino-acid sequences of the GnRH preprohormone from 4 mammalian species and the peptide sequence predicted to be produced by the G29GfsX12 mutation of patient 1. Yellow, Region processed to produce the GnRH decapeptide; gray, region that gives rise to the GAP; red, amino acids altered by G29GfsX12 (with premature stop codon indicated by an asterisk); asterisks below, amino acids altered by sequence variants in GNRH1. (C) Sequence traces indicating base-pair changes (arrows).
A homozygous frameshift mutation ([c.87delA] + [c.87delA], [p.G29GfsX12] + [p.G29GfsX12]) was identified in patient 1 (Table 1 and Fig. 3). This change lies in the codon encoding the sixth amino acid of the GnRH decapeptide and is predicted to alter all amino acids C-terminal to this residue, with premature termination after 11 aa (Fig. 3).
Four heterozygous sequence variants not seen in control subjects were found in nIHH patients (Table 1 and Fig. 3). Patient 2 has a rare variant resulting in an amino-acid substitution in the eighth residue of the GnRH decapeptide ([c.91C>T] + [=], [p.R31C] + [=]) (Table 1 and Fig. 3). The same heterozygous change in GNRH1 was found in the patient's twin daughters and niece (Fig. 2). Patient 3 carries a heterozygous rare variant ([c.217C>T] + [=], [p.R73X] + [=]) that causes premature termination in the GAP region of the GnRH precursor (Table 1 and Fig. 3). This change was also found in his sister (Fig. 2). Patient 4 is heterozygous for the variant [c.172A>T] + [=], [p.T58S] + [=], which causes an amino acid substitution in a nonconserved residue in the GAP region. Patient 5 carries a heterozygous rare variant ([c.52G>A] + [=], [p.V18M] + [=]) that alters an amino acid in the signal peptide. This amino acid change is not predicted to alter signal peptide recognition or cleavage. The patient's mother, sister, and niece also carry this variant (Fig. 2).
SNP rs6185 was seen in 135 (22%) of the 620 patient alleles and 133 (35%) of the 384 control alleles; the reported allele frequency of this SNP is 18–30% in Caucasians and 52–61% in Asians.* One patient and one control subject were found to be heterozygous for SNP rs6186, which has an allele frequency of 1–4%.*
Discussion
We have identified a homozygous frameshift mutation in GNRH1 in a patient with severe nIHH. This establishes GNRH1 as a cause of nIHH. We also identified rare heterozygous sequence variants in GNRH1 in 4 nIHH patients of 310 patients screened.
The frameshift mutation in patient 1 is unquestionably null because the C terminus of GnRH is essential for its function (reviewed in ref. 27). Patient 1's severe phenotype is consistent with complete loss of GnRH activity. His minimal rise in gonadotropins after administration of exogenous GnRH indicates lack of prior exposure to GnRH, and his history of microphallus and cryptorchidism underscores the importance of GnRH for penile growth and testicular descent during fetal development (28).
GNRH1 expression in the placenta has been well described (29), but patient 1's gestation and delivery demonstrates that zygotic GnRH is not strictly required for placental function. Indeed, GNRH1 expression has been described in a number of other tissues, including prostate, retina, and developing teeth (25, 30, 31), but the fact that patient 1 exhibits no obvious phenotypes aside from IHH argues against a critical function for GNRH1 in these tissues. GnRH is also expressed in the testes (32), and at face value patient 1's subnormal response to hCG stimulation might suggest a direct role for GnRH in the testes. However, his Leydig cell dysfunction could be a result of his cryptorchidism or his hypogonadotropism during development, because patients with severe nIHH require months of gonadotropin or pulsatile GnRH therapy to achieve Leydig cell maturation and normalization of serum testosterone (33). Thus, his poor response to hCG does not necessarily indicate a direct function of GnRH in the testes. To the contrary, his testicular biopsy did not reveal any obvious abnormalities.
The R31C change in patient 2 alters the eighth amino acid of the GnRH decapeptide. Substitutions of histidine, glutamine, leucine, serine, tyrosine, or tryptophan at this position markedly reduce the ability of GnRH to bind and activate mammalian GnRH receptors (34, 35). Lu et al. (35) propose that the human GnRH receptor has evolved to specifically recognize this arginine residue, and they have identified a residue in the human GnRH receptor (Asn7.45) that confers this specificity. Thus, the R31C change is likely to cause significant loss of GnRH function. The patient's pedigree suggests an autosomal dominant mode of inheritance, with individuals from 3 generations with nIHH. Two of the patient's daughters (DNA was not available for her eldest daughter) and her niece are all heterozygous for the R31C mutation, consistent with the possibility that the mutation is responsible for their reproductive phenotypes. However, the reproductive phenotypes in this family appear somewhat less severe, because patient 2 exhibited reversal of IHH, and the patient's niece had hypothalamic amenorrhea, with her reproductive phenotype becoming apparent only in the presence of an external stressor.
The R73X variant in patient 3 causes truncation of the GAP but is predicted to leave the GnRH decapeptide intact. It is possible that the resulting transcript is rapidly degraded, although the proximity of the premature stop codon to a splice junction may allow for escape from nonsense-mediated decay (36). Thus, the effect of this variant on GnRH synthesis is unclear. Nevertheless, this patient's pedigree suggests an autosomal dominant inheritance pattern with variable expressivity, because his father had the milder reproductive phenotype of delayed puberty. The patient's father is inferred to have carried the R73X mutation; the fact that the patient's sister carries the mutation demonstrates parental transmission, but the patient's mother does not carry the mutation.
The T58S change in patient 4 lies in a residue in the GAP region of the prohormone that is not conserved across species, and it is unclear whether this conservative amino acid substitution disrupts GnRH synthesis or function. Also, the consanguinity of his parents implies a recessive mode of inheritance of his disease. Furthermore, it is unclear how a mutation in GNRH1 could contribute to the patient's retinitis pigmentosa, although it is intriguing that GnRH-immunoreactive fibers have been observed in the mammalian retina (37).
The V18M variant in patient 5, which lies in the signal peptide, is not predicted to alter signal peptide function. Furthermore, his nIHH and adrenal insufficiency are readily attributable to his frameshift mutation in NR0B1/DAX1 (26). Nevertheless, it is notable that 3 female carriers of the NR0B1/DAX1 mutation in patient 5's family had delayed puberty (26) and are also heterozygous for the GNRH1 variant. This raises the possibility that the GNRH1 variant contributed to their reproductive phenotypes.
How might heterozygous variants in GNRH1 cause or contribute to nIHH? It is possible that our screening strategy failed to identify mutations in other regions, such as transcriptional regulatory elements, that disrupt the function of the seemingly unaffected GNRH1 allele. It is also possible that the heterozygous mutations in GNRH1 do not contribute to the pathogenesis of nIHH and that only the homozygous frameshift mutation is causal. However, there is clear precedent for association of nIHH/KS with heterozygous mutations in FGF8 and PROK2, which also encode secreted ligands (9, 10, 14, 15). Another possibility is that heterozygous GNRH1 mutations act in conjunction with mutations in other genes to cause nIHH, a mechanism that has been observed for several other nIHH/KS genes (15, 38–40). It is also possible that some mutations in GNRH1 have dominant effects. One potential mechanism is exemplified by mutations in AVP that cause autosomal dominant neurohypophyseal diabetes insipidus because of neurotoxicity of the mutant gene products (reviewed in ref. 41); mutations in GNRH1 may have similar effects. In particular, the R31C mutation in patient 2 could potentially form inappropriate disulfide bonds, resulting in misfolded peptides or aggregates that interfere with the function of the endoplasmic reticulum and cause GnRH neuronal dysfunction.
GNRH1 is an obvious candidate gene for nIHH, so why have mutations in GNRH1 not been identified to date? One possibility is that functional mutations in genes encoding ligands arise less frequently than in genes encoding receptors because of differences in size between ligands and their cognate receptors. Encoding a peptide product of only 92 aa, GNRH1 represents a smaller “target” for mutation than the 328 aa encoded by GNRHR. Indeed, for other ligand–receptor pairs implicated in nIHH/KS, fewer mutations have been reported in genes encoding ligands (FGF8 and PROK2) than in genes encoding receptors (FGFR1 and PROKR2; refs. 9, 10, and references therein). An alternative explanation for the rarity of GNRH1 mutations is that they are rapidly eliminated from the population. This could occur from inability to transmit mutations to future generations, as would be expected from mutations that cause a reduction in fertility. The expression of GNRH1 in the placenta (29) raises the possibility that mutations may cause lethality, which would also lead to elimination of mutations from the population, although again patient 1's normal gestation argues against an essential role for zygotic GnRH in the placenta.
Our study fills a long-standing gap in the genetics of nIHH. The finding of a homozygous mutation in a patient with nIHH firmly establishes mutations in GNRH1 as a rare cause of nIHH. Furthermore, the discovery of heterozygous GNRH1 changes in patients with familial nIHH suggests that mutations in GNRH1 may also act in a dominant fashion. These patients offer a rare opportunity to study the effects of human GnRH deficiency throughout the life cycle and may provide insight into functions of GnRH outside the hypothalamus.
Methods
Patients.
All studies were approved by the Institutional Review Board (IRB) of MGH, and informed consent was obtained from all patients enrolled in the study. Participants in the study were either evaluated clinically by the Reproductive Endocrine Associates of MGH or were referred directly by their physicians to participate in our genetic studies. The diagnosis of nIHH was based on the absence of spontaneous puberty and low sex steroids (testosterone, ≤3.4 nmol/L; estradiol, ≤73 pmol/L) in the setting of inappropriately normal or low gonadotropin levels and an intact sense of smell. Additional evidence for a diagnosis of nIHH was provided by the following: (i) absence of LH pulses during 12–24 h of blood sampling every 10 min (see SI Methods); (ii) normal basal and stimulated levels of thyroid-stimulating hormone, prolactin, growth hormone, and cortisol; and (iii) absence of abnormalities on imaging of the hypothalamic-pituitary region. Whenever possible, patients were interviewed by both a clinical investigator and a genetics counselor by using an IRB-approved questionnaire so that a full family pedigree could be obtained. Control subjects were also evaluated at MGH with detailed histories and physical examinations.
Of the 310 patients with nIHH screened, 212 were Caucasian, 12 African-American, 23 Asian, 1 Native American, 3 mixed race, and 59 not assessed for ethnicity. Of the 192 control subjects, 154 were Caucasian, 34 Asian, 2 Native Hawaiian/Pacific Islander, and 2 mixed race.
Sequence Analysis.
Genomic DNA was extracted from peripheral blood leukocytes or cultured white blood cells. All exons of GNRH1 and 50 bp of intronic DNA flanking each exon were sequenced by Polymorphic DNA Technologies. Sequence variants found in patients but not controls were verified by amplification of the GNRH1 coding region by PCR by using Taq polymerase (Fisher Scientific) under standard conditions with the following primers: for exon 1, 5′-CTCTGACTTCCATCTTCTGC-3′ and 5′-GCCTTATCTCACCTGGAGC-3′ (annealing temperature, 60 °C); for exon 2, 5′-CTGCAACTTTCCCAATCTCC-3′ and 5′-GAGGAGTCAGGAATGTAAGC-3′ (annealing temperature, 55 °C); and for exon 3, 5′-CTTAGCACTAACTAGAGC-3′ and 5′-GTGCAACTTGGTGTAAGG-3′ (annealing temperature, 49 °C). PCR products were purified by using the QIAquick PCR Purification kit (Qiagen) and sequenced with the same primers by the MGH DNA Sequencing Core. FGFR1 and PROKR2 were sequenced as described in refs. 42 and 43, respectively. Signal peptide recognition and cleavage were predicted by using SignalP 3.0 (44) and Sig-Pred (45).
Supplementary Material
Acknowledgments.
We thank Martin Dym for assistance with interpretation of testicular histology. This work was supported by National Institutes of Health/National Institute of Child Health and Human Development Grant U54 HD028138. Y.-M.C. received support from National Institutes of Health/National Institute of Child Health and Human Development Grant F32 HD056759.
Note Added in Proof.
Bouligand et al. (46) recently reported a homozygous frameshift mutation in two siblings with IHH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903449106/DCSupplemental.
Database of Single Nucleotide Polymorphisms (dbSNP) (National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD), www.ncbi.nlm.nih.gov/SNP/ [dbSNP accession nos. rs6185–rs6186 (dbSNP Build ID: 129)].
References
- 1.Gore AC. GnRH: The Master Molecule of Reproduction. Boston: Kluwer Academic Publishers; 2002. [Google Scholar]
- 2.Crowe S, Cushing H, Homans J. Experimental hypophysectomy. Bull Johns Hopkins Hospital. 1910;21:127–167. [Google Scholar]
- 3.Harris GW. The induction of ovulation in the rabbit by electrical stimulation of the hypothalamo-hypophysial mechanism. Proc R Soc Lond B Biol Sci. 1937;122:374–394. [Google Scholar]
- 4.Hinsey JC. The relation of the nervous system to ovulation and other phenomena of the female reproductive tract. Cold Spring Harbor Symp Quant Biol. 1937;5:269–279. [Google Scholar]
- 5.Brooks CM. A study of the mechanism whereby coitus excites the ovulation-producing activity of the rabbit's pituitary. Am J Physiol. 1938;121:157–177. [Google Scholar]
- 6.Taubenhaus M, Soskin S. Release of luteinizing hormone from the anterior hypophysis by an acetylcholine-like substance from the hypothalamic region. Endocrinology. 1941;29:958–968. [Google Scholar]
- 7.Schally AV, et al. Isolation and properties of the FSH and LH-releasing hormone. Biochem Biophys Res Comm. 1971;43:393–399. doi: 10.1016/0006-291x(71)90766-2. [DOI] [PubMed] [Google Scholar]
- 8.Amoss M, et al. Purification, amino acid composition and N-terminus of the hypothalamic luteinizing hormone releasing factor (LRF) of ovine origin. Biochem Biophys Res Commun. 1971;44:205–210. doi: 10.1016/s0006-291x(71)80179-1. [DOI] [PubMed] [Google Scholar]
- 9.Gajdos ZK, Hirschhorn JN, Palmert MR. What controls the timing of puberty? An update on progress from genetic investigation. Curr Opin Endocrinol Diabetes Obes. 2009;16:16–24. doi: 10.1097/MED.0b013e328320253c. [DOI] [PubMed] [Google Scholar]
- 10.Kim H-G, Bhagavath B, Layman LC. Clinical manifestations of impaired GnRH neuron development and function. Neurosignals. 2008;16:165–182. doi: 10.1159/000111561. [DOI] [PubMed] [Google Scholar]
- 11.Franco B, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 12.Legouis R, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 13.Dodé C, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 14.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodé C, et al. Kallmann syndrome: Mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HG, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson RS, et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 18.de Roux N, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS-1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seminara SB, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 20.de Roux N, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 21.Topaloglu AK, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2008;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattanach BM, Iddon CA, Charlton HM, Chiappa SA, Fink G. Gonadotropin-releasing hormone deficiency in a mutant mouse with hypogonadism. Nature. 1977;269:338–340. doi: 10.1038/269338a0. [DOI] [PubMed] [Google Scholar]
- 23.Mason AJ, et al. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234:1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- 24.Mason AJ, et al. The hypogonadal mouse: Reproductive functions restored by gene therapy. Science. 1986;234:1372–1378. doi: 10.1126/science.3097822. [DOI] [PubMed] [Google Scholar]
- 25.Tiong J, Locastro T, Wray S. Gonadotropin-releasing hormone-1 (GnRH-1) is involved in tooth maturation and biomineralization. Dev Dyn. 2007;236:2980–2992. doi: 10.1002/dvdy.21332. [DOI] [PubMed] [Google Scholar]
- 26.Seminara SB, Achermann JC, Genel M, Jameson JL, Crowley WF. X-linked adrenal hypoplasia congenita: A mutation in DAX1 expands the phenotypic spectrum in males and females. J Clin Endocrinol Metab. 1999;84:4501–4509. doi: 10.1210/jcem.84.12.6172. [DOI] [PubMed] [Google Scholar]
- 27.Sealfon SC, Weinstein H, Millar RP. Molecular mechanisms of ligand interaction with the gonadotropin-releasing hormone receptor. Endocr Rev. 1997;18:180–205. doi: 10.1210/edrv.18.2.0295. [DOI] [PubMed] [Google Scholar]
- 28.Grumbach MM. A window of opportunity: The diagnosis of gonadotropin deficiency in the male infant. J Clin Endocrinol Metab. 2005;90:3122–3127. doi: 10.1210/jc.2004-2465. [DOI] [PubMed] [Google Scholar]
- 29.Siler-Khodr TM, Khodr GS. Content of luteinizing hormone-releasing factor in the human placenta. Am J Obstet Gynecol. 1978;130:216–219. doi: 10.1016/0002-9378(78)90369-1. [DOI] [PubMed] [Google Scholar]
- 30.Harrison GS, Wierman ME, Nett TM, Glode LM. Gonadotropin-releasing hormone and its receptor in normal and malignant cells. Endocr Relat Cancer. 2004;11:725–748. doi: 10.1677/erc.1.00777. [DOI] [PubMed] [Google Scholar]
- 31.Cheng CK, Leung PCK. Molecular biology of gonadotropin-releasing hormone (GnRH)-I, GnRH-II, and their receptors in humans. Endocr Rev. 2005;26:283–306. doi: 10.1210/er.2003-0039. [DOI] [PubMed] [Google Scholar]
- 32.Bahk JY, et al. Stage specific identification of the expression of GnRH mRNA and localization of the GnRH receptor in mature rat and adult human testis. J Urol. 1995;154:1958–1961. [PubMed] [Google Scholar]
- 33.Pitteloud N, et al. Predictors of outcome of long-term GnRH therapy in men with idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2002;87:4128–4136. doi: 10.1210/jc.2002-020518. [DOI] [PubMed] [Google Scholar]
- 34.Millar RP, Flanagan CA, Milton RCL, King JA. Chimeric analogues of vertebrate gonadotropin-releasing hormones comprising substitutions of the variant amino acids in positions 5, 7, and 8. J Biol Chem. 1989;264:21007–21013. [PubMed] [Google Scholar]
- 35.Lu Z-L, Coetsee M, White CD, Millar RP. Structural determinants for ligand-receptor conformational selection in a peptide G protein-coupled receptor. J Biol Chem. 2007;282:17921–17929. doi: 10.1074/jbc.M610413200. [DOI] [PubMed] [Google Scholar]
- 36.Mühlemann O. Recognition of nonsense mRNA: Towards a unified model. Biochem Soc Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- 37.Wirsig-Wiechmann CR, Wiechmann AF. Vole retina is a target for gonadotropin-releasing hormone. Brain Res. 2002;950:210–217. doi: 10.1016/s0006-8993(02)03039-1. [DOI] [PubMed] [Google Scholar]
- 38.Pitteloud N, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitteloud N, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canto P, Munguía P, Söderlund D, Castro JJ, Méndez JP. Genetic analysis in patients with Kallmann syndrome: Coexistence of mutations in prokineticin receptor 2 and KAL1. J Androl. 2009;30:41–45. doi: 10.2164/jandrol.108.005314. [DOI] [PubMed] [Google Scholar]
- 41.Christensen JH, Rittig S. Familial neurohypophyseal diabetes insipidus—an update. Semin Nephrol. 2006;26:209–223. doi: 10.1016/j.semnephrol.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Pitteloud N, et al. Reversible Kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 43.Cole LW, et al. Mutations in Prokineticin 2 and Prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: Molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 45.Bradford JR. Leeds, UK: Univ of Leeds; 2001. In silico methods for prediction of signal peptides and their cleavage sites, and linear epitopes. MRes thesis. [Google Scholar]
- 46.Bouligand J, et al. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med. 2009;360:2742–2748. doi: 10.1056/NEJMoa0900136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.