Abstract
Numerous studies have implicated the pRB family of nuclear proteins in the control of cell cycle progression. Although over-expression experiments have revealed that each of these proteins, pRB, p107, and p130, can induce a G1 cell cycle arrest, mouse knockouts demonstrated distinct developmental requirements for these proteins, as well as partial functional redundancy between family members. To study the mechanism by which the closely related pRB family proteins contribute to cell cycle progression, we generated 3T3 fibroblasts derived from embryos that lack one or more of these proteins (pRB−/−, p107−/−, p130−/−, pRB−/−/p107−/−, pRB−/−/p130−/−, and p107−/−/p130−/−). By comparing the growth and cell cycle characteristics of these cells, we have observed clear differences in the manner in which they transit through the G1 and S phases as well as exit from the cell cycle. Deletion of Rb, or more than one of the family members, results in a shortening of G1 and a lengthening of S phase, as well as a reduction in growth factor requirements. In addition, the individual cell lines showed differential regulation of a subset of E2F-dependent gene promoters, as well as differences in cell cycle-dependent kinase activity. Taken together, these observations suggest that the closely related pRB family proteins affect cell cycle progression through distinct biochemical mechanisms and that their coordinated action may contribute to their diverse functions in various physiological settings.
The retinoblastoma (RB) family of proteins (pRB, p107, and p130) has been shown to regulate the activity of members of the E2F family of heterodimeric transcription factors in a variety of cell culture studies. The RB proteins bind directly to E2F proteins and repress E2F-mediated transcription, a function that is regulated by cyclin-dependent kinase (cdk) phosphorylation. Thus, regulation of the various E2F family proteins potentially links transcriptional activation and repression to the control of cell cycle progression. The activity of the E2F proteins is known to be regulated in a cell cycle-dependent manner, and fluctuations in E2F activity enable the coupling of an intricate web of gene expression programs with cell cycle position (1).
Transfection experiments have shown that over-expression of E2F or cdks promotes S phase entry, whereas over-expression of the pRB family of proteins arrests cells in the G1 phase of the cell cycle (2–4). The observation that the growth-suppressive functions of the pRB, p107, and p130 proteins map to a domain shown to mediate E2F binding (5–9) suggests that the effects of pRB family proteins on cell proliferation are mediated by their repressive effects on E2F-mediated gene expression. The repressive action of the pRB family of proteins occurs, at least in part, via the recruitment of histone deacetylase complexes to promoter regions, mediating deacetylation of histones, which results in chromatin condensation and subsequent inhibition of transcription (10–12). Transcriptional activation by E2F and/or repression by E2F-pRB family member complexes, has been examined in over-expression assays for a variety of promoters whose gene products are required for cellular events such as DNA synthesis and cell cycle regulation (1). However, in the context of over-expression, it is difficult to establish whether a particular family member is specifically required for the normal regulation of individual promoters.
The generation and characterization of mice deficient in pRB, p107, and/or p130 proteins revealed both unique and redundant functions for these proteins during development. Mice deficient in pRB die as embryos between days 13 and 15 of gestation, exhibiting defects in both differentiation and proliferation. Mice deficient in either p107 or p130 develop normally and exhibit no obvious adult phenotypes (13, 14), although genetic background differences have recently been shown to play a major role in determining the consequences of genetic loss of p107 and p130 (15, 16). Furthermore, intercrossing these mouse strains or generating chimeric animals that eliminate more than one protein has revealed significant functional overlap within this gene family (13, 17–19). The phenotypes seen in these mice potentially reflect cell cycle changes mediated by deregulation of E2F target genes, but could also be due to changes in other transcriptional programs involved in proliferation or differentiation control. Indeed, pRB has been shown to interact with and regulate the activity of a large number of diverse transcription factors.
To clarify the role of the individual pRB family members in the regulation of the cell cycle and E2F-dependent transcription, we sought to establish a genetically defined cellular system. We generated immortalized 3T3 fibroblast cell lines from mouse embryos deficient in various combinations of pRB, p107, and p130 and examined the cell cycle characteristics of these cells. Our experiments indicate that the combined activities of the pRB family members affect the mechanism by which cells transit through and exit the cell cycle. Moreover, the differences in the cell cycle properties observed among the various genotypes correlate with the regulation of E2F-sensitive promoters, suggesting that these proteins may function coordinately to couple gene transcription to cell cycle progression.
Materials and Methods
Cell Culture, Establishment of Cells, and Differentiation Protocol.
The 3T3 cells were established by using the protocol described by Todaro and Green (20). In short, cells were counted and seeded in DMEM supplemented with 10% calf serum (CS) at a density of 106 cells per 10-cm dish every 3 days with a medium change the subsequent day.
Flow Cytometric Analysis.
Asynchronously growing cells seeded at 7 × 105 cells per 10-cm dish for 24 h (Fig. 1), confluent, as well as cells maintained at confluence or under serum-deprived conditions were labeled with 10 μM BrdUrd cell labeling reagent (Amersham Pharmacia) for 30 min, harvested, and analyzed with a fluorescence-activated cell sorter (FACS) according to the manufacturer (Becton Dickinson). In short, cells were fixed in 80% ethanol, the DNA was denatured by treatment with 2 M HCl/0.5% Triton X-100 at room temperature for 30 min followed by a neutralization step in 0.1 M Na2B4O7 , pH 8.5. For immunofluorescence staining, anti-BrdUrd antibodies (Becton Dickinson) and FITC-conjugated horse anti-mouse antibodies (Vector Laboratories) were used. The immunofluorescence step is followed by an incubation in propidium iodide and RNase prior to two-dimensional FACS analysis using cellquest software (Becton Dickinson). The number of gated cells in G1 or G2/M is presented as %, and the number of gated cells incorporating BrdUrd at the time of the analysis is presented as % cells in S phase. To measure the relative cell size of the different cell lines, single cells with a DNA content ranging from 2N to 4N were gated, and forward scatter height (FSC-H) histograms were plotted. The mean FSC-H was calculated by using cellquest software (Becton Dickinson).
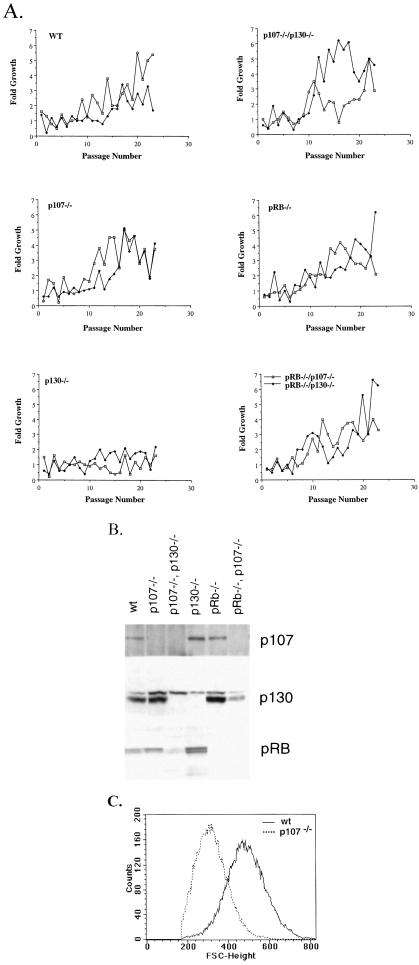
Figure 1.
Establishment of cells, protein expression, and cell size profiles of 3T3 cells deficient in pRB, p107, and/or p130. (A) Two independent isolates of MEFs, prepared from 13.5 days postcoitus embryos of mixed background (SV129/C57BL6) were used for all genotypes, except for pRB−/−/p107−/− and pRB−/−/p130−/− cells where only one isolate from an 11.5 days postcoitus embryo was used. The figure shows growth curves from these cell lines during the immortalization process. The y axis represents -fold growth in 3 days when seeded at a density of 106 cells per 10-cm dish, and the x axis is passage number (B). pRB, p107, p130 protein expression in wt 3T3 cells as well as lines deficient in p107, p130, pRB, p107/p130, and pRB/p107 at passage 45. (C) Comparison of the cell size of wt and p107-deficient 3T3 cells. Asynchronously growing wt (solid line) or p107−/− cells (dashed lines) were harvested and analyzed by FACS. FSC-H histograms, a relative measure of cell size is shown.
For the nocodazole arrest and release experiments, cells were plated at 8 × 105 cells per 10-cm dish for 24 h followed by nocodazole treatment (50 ng/ml) for 10 h, whereafter the mitotic cells were shaken off, washed, and replated at the same density. At the indicated points, the cells were labeled for 30 min with BrdUrd and analyzed as described above.
Protein Analysis and Kinase Assays.
Protein extracts from total cells were harvested from proliferating cells of different genotypes in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). Aliquots of protein samples were separated by SDS/8% PAGE (pRB, P107, p130), transferred to poly(vinylidene difluoride) membranes (Immobilon-P; from Millipore), and probed with relevant antisera. Mouse monoclonal antibodies used to detect pocket proteins were as follows: for pRB (G3-245; PharMingen), for p107 (SD9), for p130 (RB-2; Transduction Laboratories, Lexington, KY). The immunoreactive protein species were visualized by ECL detection kit. Immunoprecipitations for p107 were done in RIPA buffer, by using a mixture of antibodies (SD4, -6, -9, -15) coupled to Sepharose beads.
For the immunoprecipitation (IP)-kinase assays, two 10-cm dishes of each cell type were washed and lysed for 10 min in 2 ml Nonidet P-40 lysis buffer [50 mM Tris⋅HCl, pH 8.0/250 mM NaCl/1% Nonidet P-40/10% glycerol/0.4 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)/5 μg/ml leupeptin/5 μg/ml aprotinin/2 mM PMSF/20 mM β-glycerophosphate/2 mM NaF/2 mM Na3VO4]. For immunoprecipitations, 250 μg of total lysate was precleared with 12.5 μl of normal rabbit serum and Staphylococcus aureus Cowan (SAC) as described (35). The cleared lysates were incubated for 1 h with 10 μl of anti-CDK2 antibody (M2; Santa Cruz Biotechnology) at 4°C, 100 μl of a 10% protein A-Sepharose slurry was added, and the mixture was rotated for another 30 min. Immunoprecipitates were washed three times in lysis buffer, and 50 μl of kinase reaction mix was added to the beads (50 mM Hepes, pH 7.6/5 mM MgCl2/2.5 mM MnCl2/0.5 μg His6-pRb/1.5 mM ATP/5 μCi of [γ-32P]ATP). The kinase reactions were incubated for 30 min at 30°C and stopped with 50 μl of 2× Laemmli sample buffer, and 50 μl of this mixture was analyzed by SDS/PAGE and autoradiography. For p27 levels, 50 μg of the indicated lysates was run on SDS/12% PAGE, transferred to a Millipore poly(vinylidene difluoride) membrane, and probed with an anti-p27 antibody (Transduction Laboratories).
Stable Transfections and Transcriptional Assays.
Transfection of the 3T3 cells was performed by using calcium phosphate or Fugene (Boehringer Mannheim). Reporter constructs carrying the p107 or the B-myb promoter driving luciferase were stably integrated into the various cell lines by using a cotransfected puromycin selectable marker (pBABE). The cells were selected in DMEM/10% CS containing 2 μg/ml puromycin, pooled, and used for transcriptional analysis. Cells were harvested the day after plating (asynchronous), 2 days after serum deprivation (serum starved, 0.1% FBS), or when the cells had reached saturation at confluence (confluence). Luciferase assays (Promega) were performed according to the supplier on an equal number of cells and expressed as relative activity compared with the asynchronous activity. Results were obtained by using a luminometer from EG & G Berthold (Wallac, Gaithersburg, MD).
Results and Discussion
Establishment and Cell Cycle Characterization of pRB-, p107-, and/or p130-Deficient 3T3 Cells.
To study the biological roles of individual pRB family members, we generated a series of 3T3 fibroblast cell lines (20) from mouse embryos lacking pRB, p107, and/or p130. 3T3 cell lines, which normally express all three of these proteins, provide a relatively homogeneous and readily manipulatable system in which to dissect functional differences within this gene family. Although other mutations are known to occur during the establishment of 3T3 lines (21–23) (see below for details), to simplify presentation, we will refer to the various cell lines by the genotype of the mutant embryos from which the 3T3 lines were derived: pRB−/−, p107−/−, p130−/−, p107−/−/p130−/−, pRB−/−/p107−/−, pRB−/−/p130−/−, and wild type (wt). Mouse embryo fibroblasts of all genotypes were relatively easily established as stable lines, with the exception of p130−/− cells, which barely proliferated for 42 passages (Fig. 1A). The reason for the large number of passages required to immortalize p130−/− cells, after which they proliferate at rates similar to immortalized wt cells (Table 1, and data not shown), is not known and requires further investigation. Multiple isolates of each cell line behaved similarly in the assays described, and the genotypically distinct cells expressed the expected protein(s) (Fig. 1B, and data not shown). During the establishment of the cell lines, most genotypes (wt, p130−/−, p107−/−/p130−/−, pRB−/−, pRB−/−/p107−/−) acquired mutations in the p53 gene but the p16/p19ARF locus seemed to be unaffected (data not shown), whereas p107−/− and the pRB−/−/p130−/− cell lines lack expression of p16 and p19ARF expression (data not shown). The number of cell lines that acquired p53 mutations rather than p16/p19 deletion is similar to what has previously been published (21–23), and it is unclear at this point whether specific mutations segregate with genotype. It is interesting to note that p21−/− cells preferentially acquire p16/p19 deletions during the immortalization process (L. Stevenson, M.C., and E.H., unpublished observations), whereas wt and pRB-deficient cells preferentially mutate p53.
Table 1.
Characteristics of 3T3 fibroblast cell lines
| Genotype | Doubling time, h | % of cells in
cell cycle phases
|
Saturation density, no. of cells per 3-cm dish | Cell size, mean FSC-H | ||
|---|---|---|---|---|---|---|
| G1 | S | G2/M | ||||
| wt | 43.5 | 41 | 39 | 19 | 1.1 × 106 | 482 |
| p107−/− | 41 | 41 | 40 | 18 | 4.9 × 106 | 318 |
| p130−/− | 45 | 40 | 39 | 22 | 1.8 × 106 | 420 |
| p107−/−/p130−/− | 41 | 30 | 62 | 6 | 4.5 × 106 | 354 |
| pRB−/− | 42 | 29 | 64 | 8 | 2.7 × 106 | 367 |
| pRB−/−/p107−/− | 41 | 35 | 59 | 5 | 3.8 × 106 | 389 |
| pRB−/−/p130−/− | 41 | 31 | 61 | 7 | 4.1 × 106 | 341 |
The doubling times were determined by plating 106 cells in a 10-cm dish followed by cell counting 3 days later. The values shown are mean doubling times at passage 55–60. The saturation densities were measured by plating cells at 2 × 105 in 3-cm dishes, followed by cell counting each day until saturation was reached. The relative size of asynchronously growing cells plated at 7 × 105 per 10-cm dish 24 h prior to harvesting was measured by performing FACS analysis and plotting FSC-H histograms. The percentage of cells in the G1, S, and G2/M phases of the cell cycle in asynchronously growing populations plated at 8 × 105 cells per 10-cm dish 24 h prior to analysis. The percentage of cells in each phase of the cell cycle (G1, S, and G2/M) was measured by BrdUrd incorporation (S phase) and two-dimensional FACS analysis. The data presented are a representative example of many experiments.
Over-expression studies have suggested some functional overlap between pRB, p107, and p130 in control of the G1 phase of the cell cycle (1), and previous studies in mouse embryo fibroblasts (MEFs) indicated that pRB−/− and p107−/−/p130−/− cells exhibit a shorter G1 phase than wt cells in serum stimulation experiments (24). We were therefore surprised to find only minor differences in the doubling times of the 3T3 cells deficient in various combinations of pRB family members, and only the p130−/− cells display slightly longer cycling time (Fig. 1A, and Table 1). However, the saturation densities of the various cell lines are quite different (Table 1). This difference correlates with differences in cell size among the various cell lines as measured by FSC-H (Fig. 1C, and Table1). Previous studies had shown that pRB−/− MEFs are smaller than wt MEFs (25). Our analysis demonstrates that this size change is not unique to Rb loss, because deletion of any pRB family member results in smaller fibroblast cells (Table 1, and Fig. 1C). In general, the cell lines that spend less time in G1 are smaller. This finding is consistent with the notion that cells deficient in pRB or more than one family member spend less time in G1 (for details see below) and are subsequently smaller than wt cells (Table 1, Fig. 2). However, p107−/− and wt cells have similar cell cycle characteristics, but p107−/− cells are the smallest of all of the cell lines (Fig. 1C). Therefore, the relative size of these cell lines does not strictly correlate with time spent in G1, suggesting that other factors contribute to overall cell size.
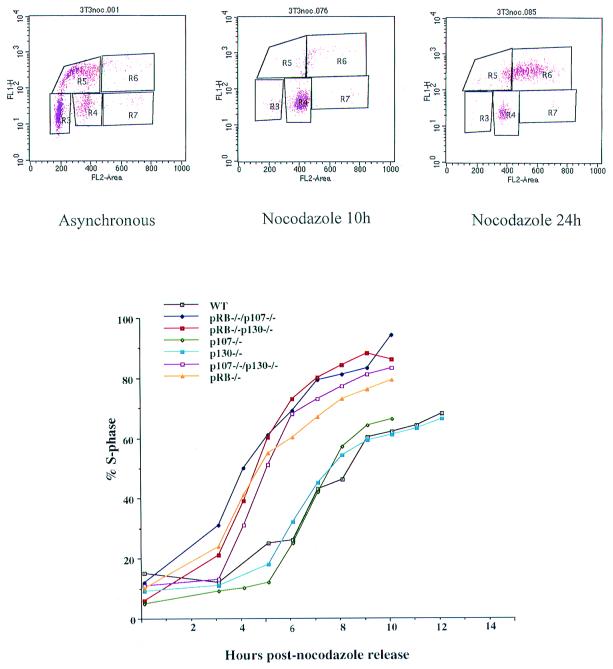
Figure 2.
Timing of the cell cycle in cells deficient in pRB, p107, and/or p130. Asynchronous cells were treated with nocodazole for 10 h, and mitotic cells were shaken off and replated. (Upper) Example of the direct FACS data (R3 = G1, R5 = S phase, and R4 = G2/M); these 3T3 cells enter endo-reduplication upon prolonged nocodazole treatment. (Lower) The timing of S phase entry measured by BrdUrd incorporation and two-dimensional FACS analysis. The y-axis represents % S phase cells and the x-axis, hours postrelease. A representative example of three experiments is shown.
Loss of RB Family Proteins Shortens G1 but Lengthens S Phase.
To investigate the cell cycle characteristics of these 3T3 cell lines in more detail, we analyzed the cell cycle profiles of asynchronously growing cell populations (Table 1). The various cell lines were plated at equivalent densities 24 h before the analysis of their cell cycle profiles, which was performed by using BrdUrd incorporation and two-dimensional FACS analysis. These experiments revealed that cells deficient in pRB, p107/p130, pRB/p107, and pRB/p130 have an increased S phase population compared with wt, p107−/−, and p130−/− cells. The larger proportion of cells in S phase could potentially be due to a shorter G1 phase and/or a lengthening of S phase because the doubling time of these cells is approximately the same. In both yeast and mammalian cells, a shortening of one phase of the cell cycle has been shown to result in a compensatory lengthening of other phases of the cell cycle, which allows the populations to maintain a relatively constant cell size (26–28). However, the mechanism by which such compensation occurs is unknown.
To distinguish between these possibilities, we monitored the cell cycle progression of a synchronized population of cells. The cells were synchronized by nocodazole arrest, followed by a mitotic shake-off and replating in the absence of drug. Using this protocol, we isolated uniform populations of metaphase-arrested cells that reentered the cell cycle in a synchronous manner (29, 30). In our initial experiments, we found that all of these cells undergo endo-reduplication of DNA if they are exposed to nocodazole for 24 h (Fig. 2 Upper Right). However, treatment of asynchronously growing cells (Fig. 2 Upper Left) with nocodazole for only 10 h gives rise to a uniform population of cells with a 4N DNA content and little evidence of endo-reduplication (Fig. 2 Upper Middle). Therefore, cells were treated with nocodazole for 10 h before the mitotic cells were shaken from the dish, washed, and replated. To measure the rate at which these cells entered S phase, cells were allowed to incorporate BrdUrd at various times after replating.
As shown in Fig. 2, pRB−/−, p107−/−/p130−/−, pRB−/−/p107−/−, and pRB−/−/p130−/− cells spend less time in G1 relative to the wt, p107−/−, and p130−/− cells. Even though all of the “double-deficient” cells enter S phase prematurely, they spend a longer time period synthesizing DNA and they enter G2 with kinetics similar to wt cells (data not shown). Thus, these results indicate that the shortening of G1 is “compensated” for by a lengthening of S phase, and appears to account for the fact that all of the cell lines exhibit a similar doubling time (Fig. 1A). A potential explanation for this observation is that derepression of a subset of E2F target genes shortens the length of G1, but other factors required for efficient synthesis of DNA are still limiting, resulting in a longer S phase. Taken together, these experiments suggest that deregulation of E2F target genes, resulting from loss of pRB, p107, and/or p130 results in earlier entry into S phase, but these cells synthesize DNA more slowly because of the absence of other rate-limiting factors required for S phase, such as cdk activity.
Derepression of E2F Target Genes Can Be Separated from Cell Cycle Progression.
Because each of the RB family proteins has been found to regulate E2F activity, we next wanted to examine potential differences in the regulation of E2F-sensitive promoters among cells of the various genotypes. Initially, we examined E2F target gene expression in nocodazole-arrested/released cells progressing through the G1 phase of the cell cycle. However, under these circumstances, it was difficult to distinguish between cell cycle-dependent and RB family-dependent changes in E2F-mediated transcriptional control. For example, we observed an earlier induction of E2F-mediated transcription in pRB−/− cells and all of the double-negative cells when compared with wt cells (data not shown), but these changes could simply be due to the fact that these cells enter S phase earlier. Therefore, to separate cell cycle-dependent changes in E2F activity from effects that are dependent on RB family proteins, but are cell cycle-independent, we performed studies of E2F target gene expression in the context of growth-arrested cells.
Initially, we examined the ability of 3T3 cells deficient in pRB family members to arrest in response to serum withdrawal or confluence. These experiments show that the various cell lines vary substantially in their ability to arrest in low serum (Fig. 3A). Thus, following serum deprivation, all of the double-negative cells exhibit a substantial fraction of BrdUrd-incorporating cells, with the pRB−/−/p130−/− cells having the highest fraction of cells in S phase, followed by pRB−/−, pRB−/−/p107−/−, and p107−/−/p130−/−. Cells deficient in p107 and p130 exhibit an intermediate phenotype compared with wt cells, which arrest well in response to serum deprivation. In sharp contrast, all of the cell lines efficiently arrest in G1 after contact inhibition, with barely detectable S phase populations (Fig. 3A).
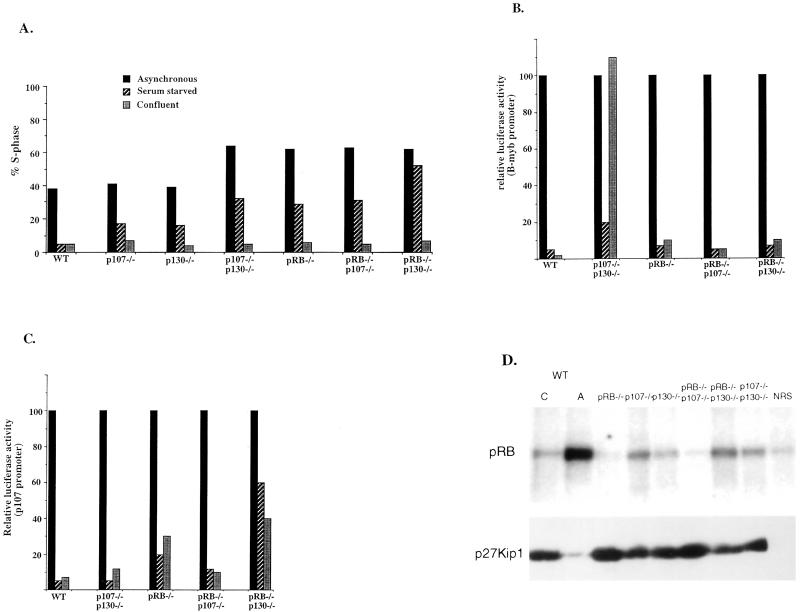
Figure 3.
Cell cycle arrest and derepression of E2F target genes in pRB, p107, and/or p130 deficient cells. (A) The % of cells in S phase, as measured by two-dimensional FACS in asynchronous (solid bars), serum-starved (striped bars), and confluent cells (gray bars) in the genotypically different cells, as indicated in the figure. (B) Relative luciferase activity from pools of cells harboring an integrated B-myb reporter under serum-starved (striped bars) and confluent conditions (gray bars) as compared with asynchronous populations (arbitrarily set to 100). A representative example from three different experiments is shown. (C) Relative luciferase activity from pools of cells harboring an integrated p107 promoter construct under serum-starved (striped bars) and confluent conditions (gray bars) as compared with asynchronous populations (arbitrarily set to 100). A representative example from three different experiments is shown. (D) (Upper) cdk2 kinase activity, using a His-tagged RB protein as a substrate in confluent (C) and asynchronous (A) wt cells and confluent knockout cell lines. (Lower) The corresponding levels of p27.
Several E2F-responsive genes had been identified whose expression appear to be deregulated in cells deficient in RB family proteins (24, 31). We chose two of these, B-myb and p107, for detailed analysis. To closely mimic physiological conditions for gene regulation, constructs harboring these promoters linked to a luciferase reporter were stably integrated into each of the 3T3 cell lines. Promoter activity was then measured in asynchronously growing and confluence-arrested cells. In addition, promoter activity was measured in serum-starved cells for comparison, because these two promoters had previously been found deregulated in serum-starved p107−/−/p130−/− (B-myb promoter) and pRB−/− (p107 promoter) cells, respectively (24). As shown in Fig. 3B, expression from the integrated B-myb reporter construct is repressed in contact inhibited wt cells. However, in p107−/−/p130−/− cells, this reduction in B-myb expression is virtually absent following confluence arrest, as was previously reported for serum-starved p107−/−/p130−/− MEFS. This derepression is specific to the combined loss of p107 and p130 because none of the other cells exhibits any significant increase in expression from this promoter when compared with wt cells. These experiments indicate that p107 and p130 are redundant in their repressive effects on the B-myb promoter under these conditions, and that the derepression observed in serum-starved cells was not simply due to their poor arrest.
Expression from the p107 promoter is also significantly repressed under confluence arrest conditions in wt cells (Fig. 3C), and this repression is also seen in p107−/−/p130−/− and pRB−/−/p107−/− cells. However, expression of the p107 promoter construct in pRB−/−, and especially pRB−/−/p130−/− cells, is derepressed under arrested conditions. This experiment suggests that not only pRB, as previously shown, but pRB in cooperation with p130 mediates repression of this promoter during confluence arrest. A similar derepression is seen upon serum starvation, however, as described earlier, in that context it is difficult to separate cell cycle position from E2F-mediated transcription (Fig. 3 B and C). Taken together, these results suggest that the various E2F-responsive promoters are differentially regulated by the combinatorial actions of the RB family members.
It is possible, based on the results presented above, that the reduced arrest of some of these cell lines following serum starvation results from the derepression of a subset of E2F target genes. This would be consistent with earlier studies suggesting that serum-arrested cells can be induced to re-enter the cell cycle by E2F over-expression (32). However, if derepression of the same set of E2F target genes occurs during confluence mediated arrest, as our analysis of a limited number of target genes suggest, this is apparently not sufficient to promote S phase entry in this context, because all of the pRB family member negative cells arrest in response to contact inhibition. There are at least two possible explanations for why these cells appear to arrest normally upon contact inhibition, but poorly upon serum deprivation. One possibility is that different sets of E2F target genes are derepressed under the two different growth conditions. However, this hypothesis will be difficult to study until all of the relevant E2F targets are identified. Alternatively, there may be a distinct type of S phase inhibitor in contact-inhibited cells that is independent of E2F activity.
It has previously been demonstrated that cdk2 kinase activity is crucial for S phase entry and that the cdk2 inhibitor p27 is up-regulated under confluent conditions (33). Furthermore, it has been shown that cyclin E, which is one of the cyclin partners for cdk2, can overcome a cell cycle block imposed by a dominant negative DP-1 construct or a nonphosphorylatable RB (34). These experiments suggest that activation of cdk2 can promote S phase entry in the absence of E2F activity. Hence, we examined the various lines for the status of p27 and the activity of cdk2 in confluence arrested cells. These experiments revealed that the level of p27 protein is increased in all of the cell lines at confluence (Fig. 3D, Lower). Moreover, the up-regulation of p27 results in a significantly lower cdk2 activity (Fig. 3D, Upper) in all of the confluent cell populations compared with wt cells, suggesting that these cells arrest because of limiting amounts of cdk2, even if E2F activity is significantly increased.
In summary, we have established a cell system in which the functions of pRB, p107, and p130 can be compared and contrasted under conditions where these proteins are not over-expressed. Our experiments suggest that loss of pRB or more than one family member results in a shortening of G1 and an associated lengthening of S phase. Furthermore, the cells deficient in pRB family members display a reduced growth factor requirement, but, independent of genotype, all of the cells arrest at confluence despite deregulation of genotype-specific E2F target genes. Taken together, these results suggest that the pRB proteins can function in a combinatorial way to regulate the expression pattern of E2F-responsive genes and the characteristics of cell cycle progression and arrest.
Acknowledgments
We thank the Weinberg, Jacks, Dyson, and Harlow laboratories for generating the knockout mice, R. Hurford and N. Dyson for establishing MEFs from these animals, and R. J. Watson and L. S. Chang for B-myb and p107 promoter constructs. We also express our sincere gratitude to N. Dyson, J. Settleman, and J. Zhao, for reading of the manuscript and all of the members of the Harlow and Dyson Laboratories for helpful discussions. M.C. has been sponsored by a MBRC Tosteson Award and a fellowship from the Medical Foundation, and S.R.S. by a fellowship from Jane Coffin Child Memorial Fund for Medical Research. This work was supported by grants from the National Institute of Health (to E.H.).
Abbreviations
- RB
retinoblastoma
- cdk
cyclin-dependent kinase
- FSC-H
forward scatter height
- wt
wild type
- MEF
mouse embryo fibroblast
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190343497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190343497
References
- 1.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 3.Lukas J, Parry D, Aagard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 4.Qin X Q, Chittenden T, Livingston D M, Kaelin W G., Jr Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 5.Nevins J R. Curr Opin Genet Dev. 1994;4:130–134. doi: 10.1016/0959-437x(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 6.Dyson N, Buchkovich K, Whyte P, Harlow E. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 7.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. Cell. 1989;58:257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 8.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Nature (London) 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 9.Whyte P, Williamson N M, Harlow E. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 10.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 11.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 12.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 13.Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 14.Lipinski M M, Jacks T. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- 15.LeCouter J E, Kablar B, Whyte P F, Ying C, Rudnicki M A. Development (Cambridge, UK) 1998;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- 16.LeCouter J E, Kablar B, Hardy W R, Ying C, Megeney L A, May L L, Rudnicki M A. Mol Cell Biol. 1998;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robanus-Maandag E, Dekker M, van der Valk M, Carrozza M-L, Jeanny J-C, Dannenberg J-H, Berns A, te Riele H. Genes Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M-H, Williams B O, Mulligan G, Mukai S, Bronson R T, Dyson N, Harlow E, Jacks T. Genes Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T. Annu Rev Genet. 1996;30:603–636. doi: 10.1146/annurev.genet.30.1.603. [DOI] [PubMed] [Google Scholar]
- 20.Todaro G J, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano M, Lee H, Chin L, Gordon-Caro C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 22.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashum R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 23.Carnero A, Hudson J D, Price C M, Beach D H. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- 24.Hurford R, Cobrinik D, Lee M-H, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 25.Herrera R E, Sah V P, Williams B O, Makela T P, Weinberg R A, Jacks T. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee A B, Dubrow R, Hamlin J L, Kletzien R F. Annu Rev Biochem. 1978;47:715–750. doi: 10.1146/annurev.bi.47.070178.003435. [DOI] [PubMed] [Google Scholar]
- 27.Prescott D M. Adv Genet. 1976;18:99–177. doi: 10.1016/s0065-2660(08)60438-1. [DOI] [PubMed] [Google Scholar]
- 28.Pringle J R, Hartwell L H. In: The Saccharomyces cerevisiae Cell Cycle. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 97–142. [Google Scholar]
- 29.Moria A O, Draetta G, Beach D, Wang J Y J. Cell. 1989;58:193–203. doi: 10.1016/0092-8674(89)90415-7. [DOI] [PubMed] [Google Scholar]
- 30.Zieve G W, Turnbull D, Mullins J M, McIntosh J R. Exp Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 31.Mulligan, G. J., Wong, J. & Jacks, T. (1998) Mol. Cell. Biol.18. [DOI] [PMC free article] [PubMed]
- 32.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 33.Polyak K, Kato J Y, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 34.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 35.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]