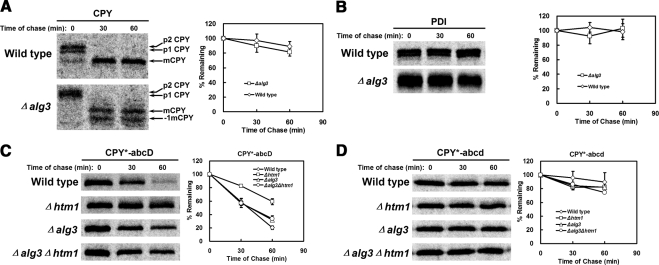
Figure 3.
Glycan structure alone is not sufficient for ERAD substrate recognition. (A and B) Wild-type and Δalg3 cells were pulse-labeled for 10 min and chased for times indicated. Proteins were immunoprecipitated from detergent lysates using polyclonal antisera and resolved by SDS-PAGE. Protein turnover was quantified by phosphorimager analysis. (A) Relative CPY turnover is plotted with representative phosphorimages shown. The Δalg3 mutant exhibits minor underglycosylation of proteins (Jakob et al., 1998). The positions of mature CPY bearing four glycans (mCPY) and three glycans (−1 mCPY) are indicated. (B) PDI stability was analyzed as described in A. (C and D) Turnover of CPY* variants were compared in wild-type, Δhtm1, Δalg3, and Δalg3Δhtm1 strains by metabolic pulse-chase analysis as in A and B. (C) Degradation of CPY*-abcD and (D) the nonglycosylated variant CPY*-abcd. The data plotted reflect three independent experiments and the SEM.